⚠️ 以下所有内容总结都来自于 大语言模型的能力,如有错误,仅供参考,谨慎使用
🔴 请注意:千万不要用于严肃的学术场景,只能用于论文阅读前的初筛!
💗 如果您觉得我们的项目对您有帮助 ChatPaperFree ,还请您给我们一些鼓励!⭐️ HuggingFace免费体验
2025-01-16 更新
Continual Deep Active Learning for Medical Imaging: Replay-Base Architecture for Context Adaptation
Authors:Rui Daniel, M. Rita Verdelho, Catarina Barata, Carlos Santiago
Deep Learning for medical imaging faces challenges in adapting and generalizing to new contexts. Additionally, it often lacks sufficient labeled data for specific tasks requiring significant annotation effort. Continual Learning (CL) tackles adaptability and generalizability by enabling lifelong learning from a data stream while mitigating forgetting of previously learned knowledge. Active Learning (AL) reduces the number of required annotations for effective training. This work explores both approaches (CAL) to develop a novel framework for robust medical image analysis. Based on the automatic recognition of shifts in image characteristics, Replay-Base Architecture for Context Adaptation (RBACA) employs a CL rehearsal method to continually learn from diverse contexts, and an AL component to select the most informative instances for annotation. A novel approach to evaluate CAL methods is established using a defined metric denominated IL-Score, which allows for the simultaneous assessment of transfer learning, forgetting, and final model performance. We show that RBACA works in domain and class-incremental learning scenarios, by assessing its IL-Score on the segmentation and diagnosis of cardiac images. The results show that RBACA outperforms a baseline framework without CAL, and a state-of-the-art CAL method across various memory sizes and annotation budgets. Our code is available in https://github.com/RuiDaniel/RBACA .
在医学成像领域,深度学习在适应和泛化到新环境时面临挑战。此外,它常常缺乏针对特定任务的充足标记数据,需要大量注释工作。持续学习(CL)通过实现从数据流中的终身学习并减轻对先前学习知识的遗忘,来解决适应性和泛化能力问题。主动学习(AL)减少了进行有效训练所需的注释数量。这项工作探讨了这两种方法(CAL),以开发用于稳健医学图像分析的全新框架。基于图像特性变化的自动识别,用于上下文适应的回放基础架构(RBACA)采用一种CL复习方法以实现从多种上下文中的持续学习,并采用AL组件来选择最具有信息性的实例进行注释。使用定义的度量标准IL-Score建立了一种评估CAL方法的新方法,该标准可以同时评估迁移学习、遗忘和最终模型性能。我们通过评估心脏图像分割和诊断的IL-Score,证明了RBACA在领域和类增量学习场景中的表现。结果表明,RBACA在各种内存大小和注释预算方面优于无CAL的基线框架以及最先进的CAL方法。我们的代码可在https://github.com/RuiDaniel/RBACA中找到。
论文及项目相关链接
Summary
本摘要探讨深度学习在医学影像领域的挑战,包括适应新语境和泛化能力的问题,以及缺乏充足标注数据的问题。文章提出了结合持续学习(CL)和主动学习(AL)的解决方案,旨在建立一个稳健的医学影像分析框架。通过采用基于图像特性变化的自动识别技术,框架实现了持续学习的适应能力和主动学习机制。实验结果显示,该框架在域和类别增量学习场景中表现出较好的效果。随着各种记忆大小和标注预算的评估,该框架优于基线框架和当前先进的CAL方法。相关代码可在链接中找到:https://github.com/RuiDaniel/RBACA。
Key Takeaways
- 深度学习在医学影像领域面临适应新语境和泛化能力的问题,以及标注数据不足的问题。
- 持续学习(CL)和主动学习(AL)被结合来解决这些问题,建立一个稳健的医学影像分析框架。
- 框架采用基于图像特性变化的自动识别的持续学习适应能力。
- 通过主动学习机制,框架能够选择最具代表性的实例进行标注,减少标注工作量。
- 实验结果显示该框架在域和类别增量学习场景中表现优越。
- 该框架在各种记忆大小和标注预算下的性能优于基线框架和当前先进的CAL方法。
点此查看论文截图
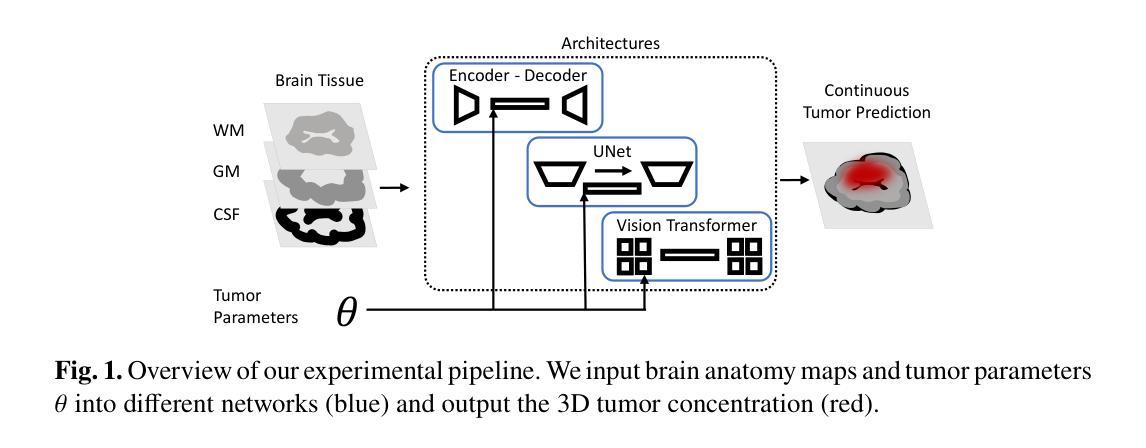
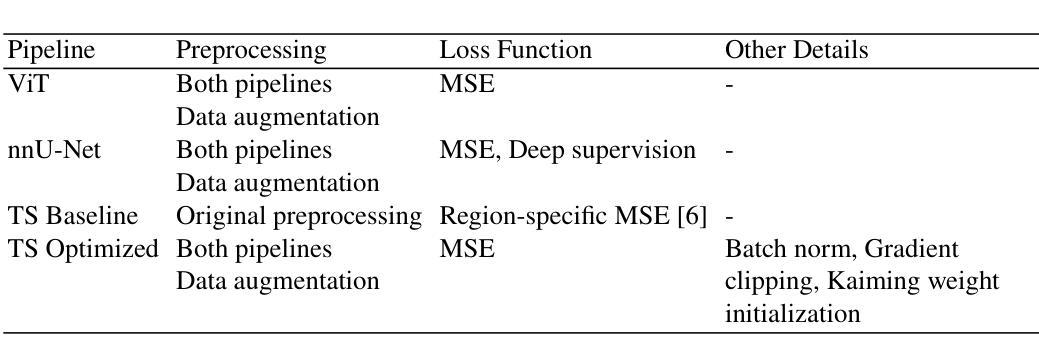
Efficient Deep Learning-based Forward Solvers for Brain Tumor Growth Models
Authors:Zeineb Haouari, Jonas Weidner, Ivan Ezhov, Aswathi Varma, Daniel Rueckert, Bjoern Menze, Benedikt Wiestler
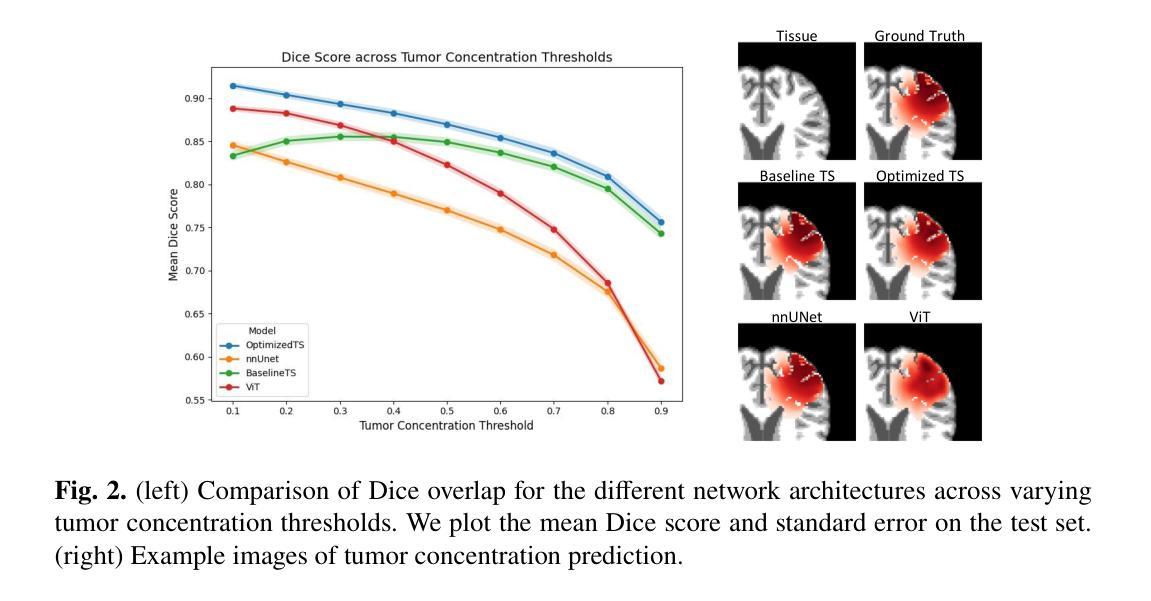
Glioblastoma, a highly aggressive brain tumor, poses major challenges due to its poor prognosis and high morbidity rates. Partial differential equation-based models offer promising potential to enhance therapeutic outcomes by simulating patient-specific tumor behavior for improved radiotherapy planning. However, model calibration remains a bottleneck due to the high computational demands of optimization methods like Monte Carlo sampling and evolutionary algorithms. To address this, we recently introduced an approach leveraging a neural forward solver with gradient-based optimization to significantly reduce calibration time. This approach requires a highly accurate and fully differentiable forward model. We investigate multiple architectures, including (i) an enhanced TumorSurrogate, (ii) a modified nnU-Net, and (iii) a 3D Vision Transformer (ViT). The optimized TumorSurrogate achieved the best overall results, excelling in both tumor outline matching and voxel-level prediction of tumor cell concentration. It halved the MSE relative to the baseline model and achieved the highest Dice score across all tumor cell concentration thresholds. Our study demonstrates significant enhancement in forward solver performance and outlines important future research directions.
胶质母细胞瘤是一种高度侵袭性的脑肿瘤,由于其预后不良和高发病率,带来了重大挑战。基于偏微分方程模型的模拟患者特异性肿瘤行为,在提高放疗计划方面,显示出提高治疗效果的广阔前景。然而,模型校准仍是瓶颈,因为优化方法(如蒙特卡洛采样和进化算法)的计算需求很高。针对这一问题,我们最近引入了一种利用基于梯度的神经正向求解器的方法,显著减少了校准时间。这种方法需要一个高度精确和完全可微分的正向模型。我们调查了多种架构,包括(i)增强的TumorSurrogate,(ii)改进的nnU-Net,(iii)3D Vision Transformer(ViT)。优化后的TumorSurrogate取得了最佳的整体结果,在肿瘤轮廓匹配和肿瘤细胞浓度的体素级预测方面都表现出色。与基线模型相比,它使均方误差减半,并在所有肿瘤细胞浓度阈值上取得了最高的Dice得分。我们的研究展示了正向求解器性能的显著提高,并概述了未来重要的研究方向。
论文及项目相关链接
Summary
胶质母细胞瘤是一种高度侵袭性的脑肿瘤,预后不良且发病率高,面临巨大挑战。基于偏微分方程模型的优化放疗计划对患者特异性肿瘤行为进行模拟,具有提高治疗效果的潜力。然而,模型校准因蒙特卡罗采样和进化算法等优化方法的高计算需求而成为瓶颈。最近,我们采用神经网络前向求解器与基于梯度的优化方法,显著减少了校准时间。该方法需要高度精确和完全可区分的前向模型。我们调查了多种架构,包括增强的TumorSurrogate、改进的nnU-Net和3D Vision Transformer(ViT)。优化后的TumorSurrogate取得最佳结果,在肿瘤轮廓匹配和肿瘤细胞浓度体素级预测方面都表现出色,相对基准模型降低了均方误差,并在所有肿瘤细胞浓度阈值上获得最高Dice得分。本研究显著提高了前向求解器的性能,并指出了重要的未来研究方向。
Key Takeaways
- 胶质母细胞瘤是一种具有高度侵袭性和不良预后的脑肿瘤。
- 基于偏微分方程模型的模拟能够优化放疗计划以提高治疗效果。
- 模型校准是一个挑战,因优化方法的高计算需求而成为瓶颈。
- 采用神经网络前向求解器与基于梯度的优化方法显著减少了模型校准时间。
- 研究了多种模型架构,包括TumorSurrogate、nnU-Net和ViT。
- 优化后的TumorSurrogate在肿瘤轮廓匹配和肿瘤细胞浓度预测方面表现最佳。
点此查看论文截图

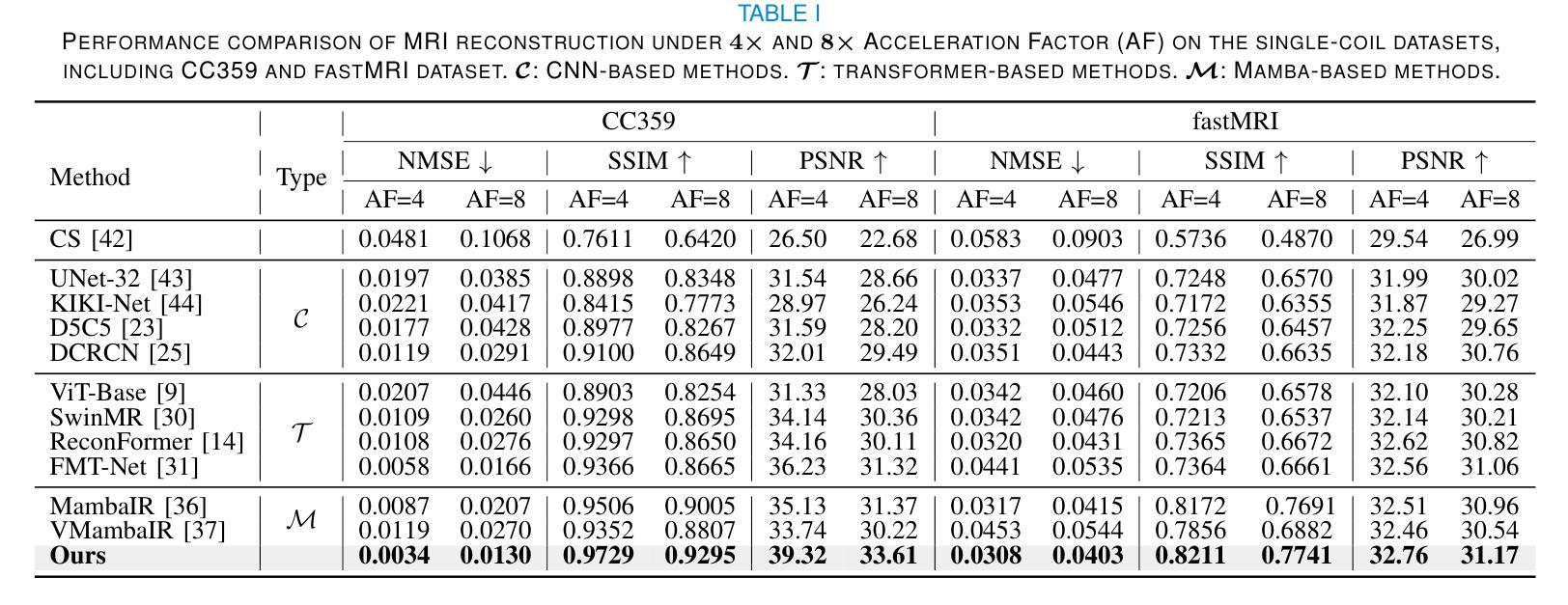
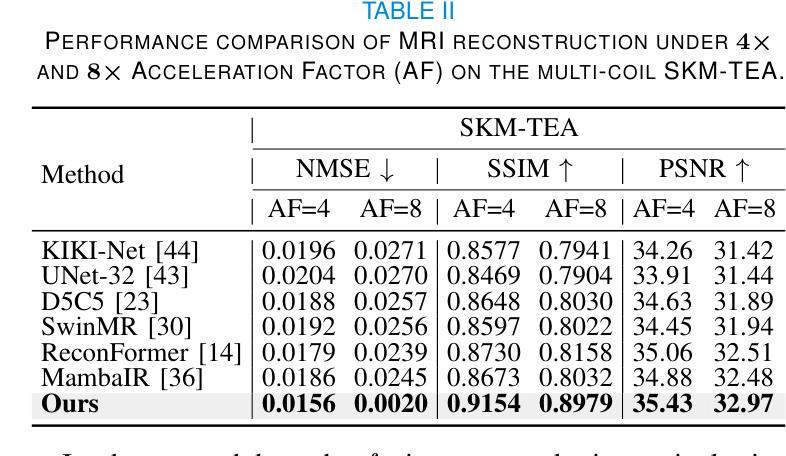
DM-Mamba: Dual-domain Multi-scale Mamba for MRI reconstruction
Authors:Yucong Meng, Zhiwei Yang, Zhijian Song, Yonghong Shi
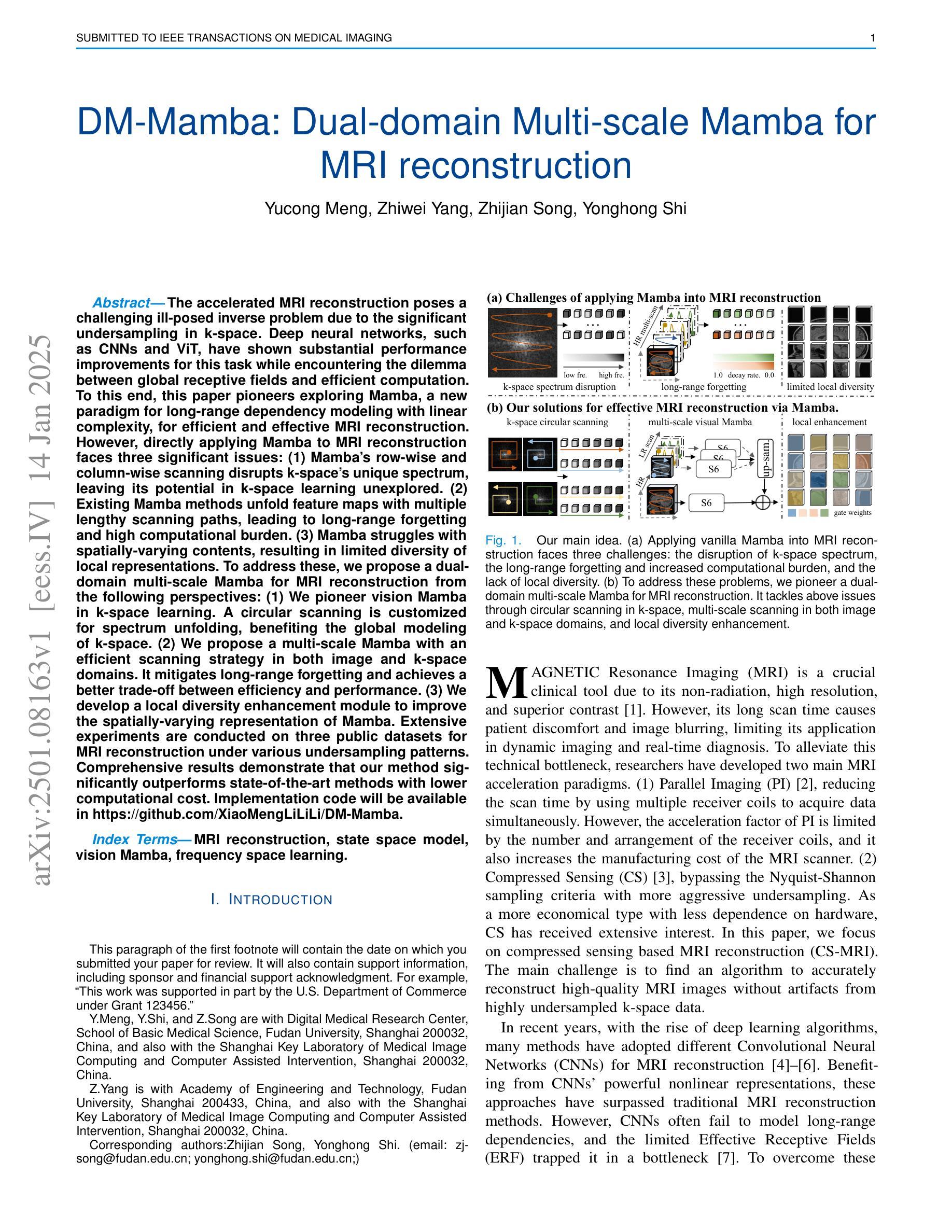
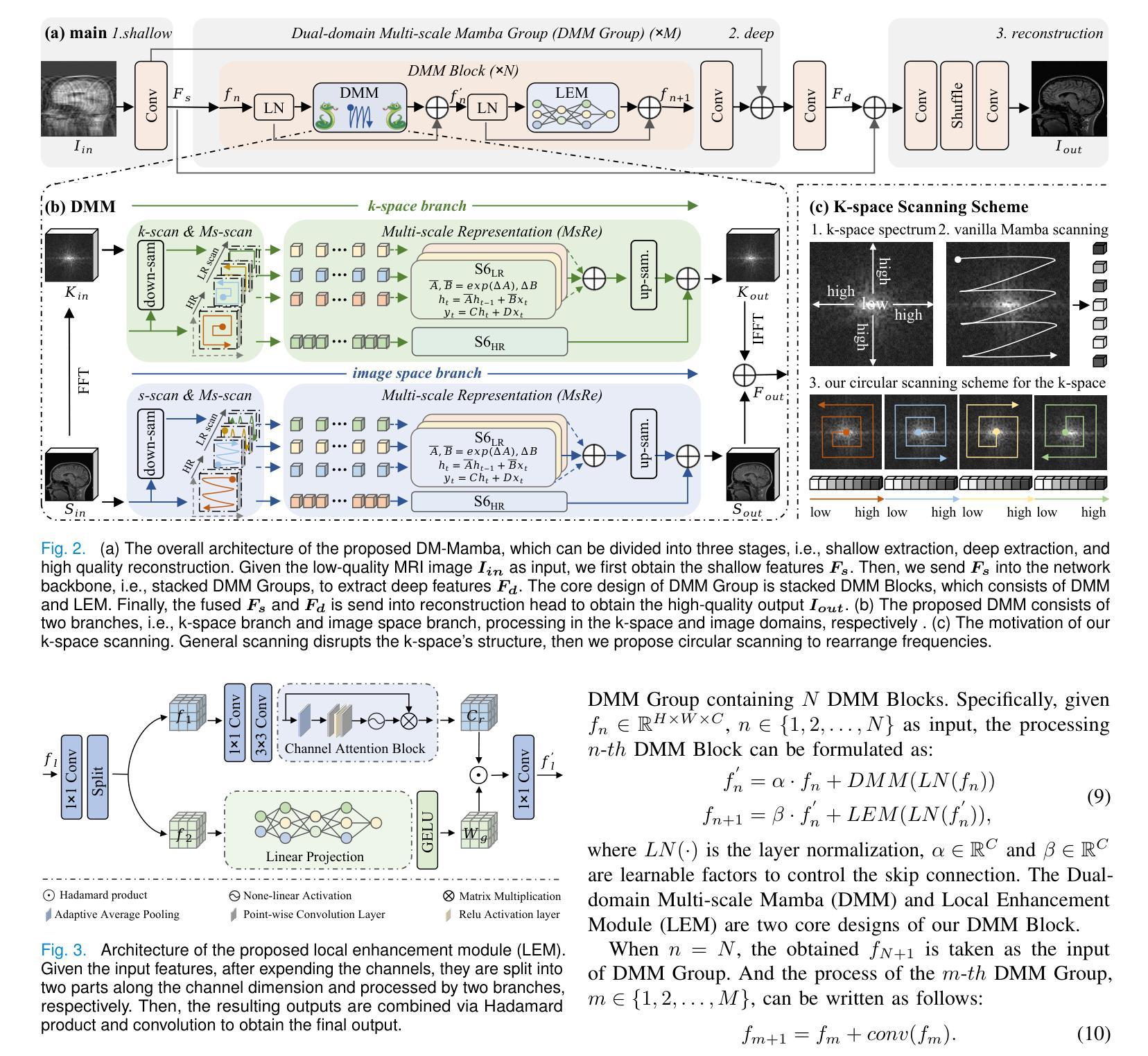
The accelerated MRI reconstruction poses a challenging ill-posed inverse problem due to the significant undersampling in k-space. Deep neural networks, such as CNNs and ViT, have shown substantial performance improvements for this task while encountering the dilemma between global receptive fields and efficient computation. To this end, this paper pioneers exploring Mamba, a new paradigm for long-range dependency modeling with linear complexity, for efficient and effective MRI reconstruction. However, directly applying Mamba to MRI reconstruction faces three significant issues: (1) Mamba’s row-wise and column-wise scanning disrupts k-space’s unique spectrum, leaving its potential in k-space learning unexplored. (2) Existing Mamba methods unfold feature maps with multiple lengthy scanning paths, leading to long-range forgetting and high computational burden. (3) Mamba struggles with spatially-varying contents, resulting in limited diversity of local representations. To address these, we propose a dual-domain multi-scale Mamba for MRI reconstruction from the following perspectives: (1) We pioneer vision Mamba in k-space learning. A circular scanning is customized for spectrum unfolding, benefiting the global modeling of k-space. (2) We propose a multi-scale Mamba with an efficient scanning strategy in both image and k-space domains. It mitigates long-range forgetting and achieves a better trade-off between efficiency and performance. (3) We develop a local diversity enhancement module to improve the spatially-varying representation of Mamba. Extensive experiments are conducted on three public datasets for MRI reconstruction under various undersampling patterns. Comprehensive results demonstrate that our method significantly outperforms state-of-the-art methods with lower computational cost. Implementation code will be available at https://github.com/XiaoMengLiLiLi/DM-Mamba.
加速MRI重建是一个具有挑战性的不适定反问题,这主要是由于k空间中的显著欠采样造成的。深度神经网络,如卷积神经网络和Vision Transformer,虽然在此任务中取得了显著的性能提升,但在全局感受野和高效计算之间遇到了困境。为此,本文率先探索Mamba这一具有线性复杂度的长距离依赖建模新范式,以实现高效且有效的MRI重建。然而,直接将Mamba应用于MRI重建面临三个主要问题:(1)Mamba的行和列扫描破坏了k空间的独特频谱,使得其在k空间学习中的潜力尚未被探索。(2)现有的Mamba方法在展开特征映射时采用了多条冗长的扫描路径,导致长距离遗忘和高计算负担。(3)Mamba在空间变化内容上遇到困难,导致局部表示多样性有限。为了解决这些问题,我们从以下角度提出了用于MRI重建的双域多尺度Mamba:(1)我们率先在k空间学习中引入视觉Mamba。为频谱展开定制了圆形扫描,有利于k空间的全局建模。(2)我们提出了一个在图像和k空间域中具有高效扫描策略的多尺度Mamba。它减轻了长距离遗忘问题,并在效率和性能之间取得了更好的平衡。(3)我们开发了一个局部多样性增强模块,以提高Mamba的空间变化表示。在三个公共数据集上进行了广泛的MRI重建实验,实验结果表明,我们的方法在较低的计算成本下显著优于最先进的方法。相关实现代码将在https://github.com/XiaoMengLiLiLi/DM-Mamba上发布。
论文及项目相关链接
Summary
此文本介绍了磁共振成像(MRI)重建的挑战性问题,以及针对该问题采用的新型模型Mamba的优缺点。为改进Mamba在MRI重建中的应用,提出了双域多尺度Mamba方法,在图像和k空间域进行高效扫描,增强局部多样性,并在三个公共数据集上进行了广泛实验验证,结果显示该方法在降低计算成本的同时显著优于现有技术。
Key Takeaways
- 磁共振成像(MRI)重建面临因k空间中显著欠采样导致的挑战性逆问题。
- Mamba模型作为长距离依赖建模的新范式,具有高效和有效的MRI重建潜力。
- 直接应用Mamba于MRI重建面临三个主要问题:对k空间独特谱的干扰、长距离遗忘和计算负担高、以及处理空间变化内容时的局限性。
- 为解决这些问题,提出了双域多尺度Mamba方法,包括在k空间进行视觉Mamba的开创性研究、采用多尺度Mamba进行图像和k空间域的高效扫描策略以及开发局部多样性增强模块。
点此查看论文截图

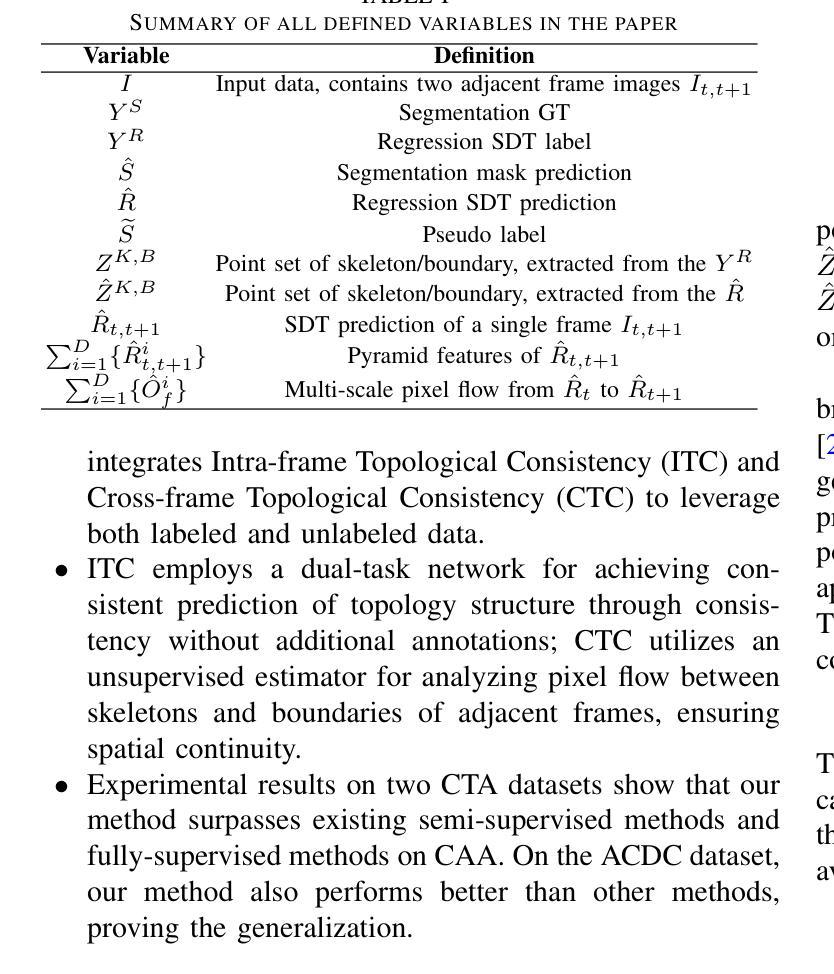
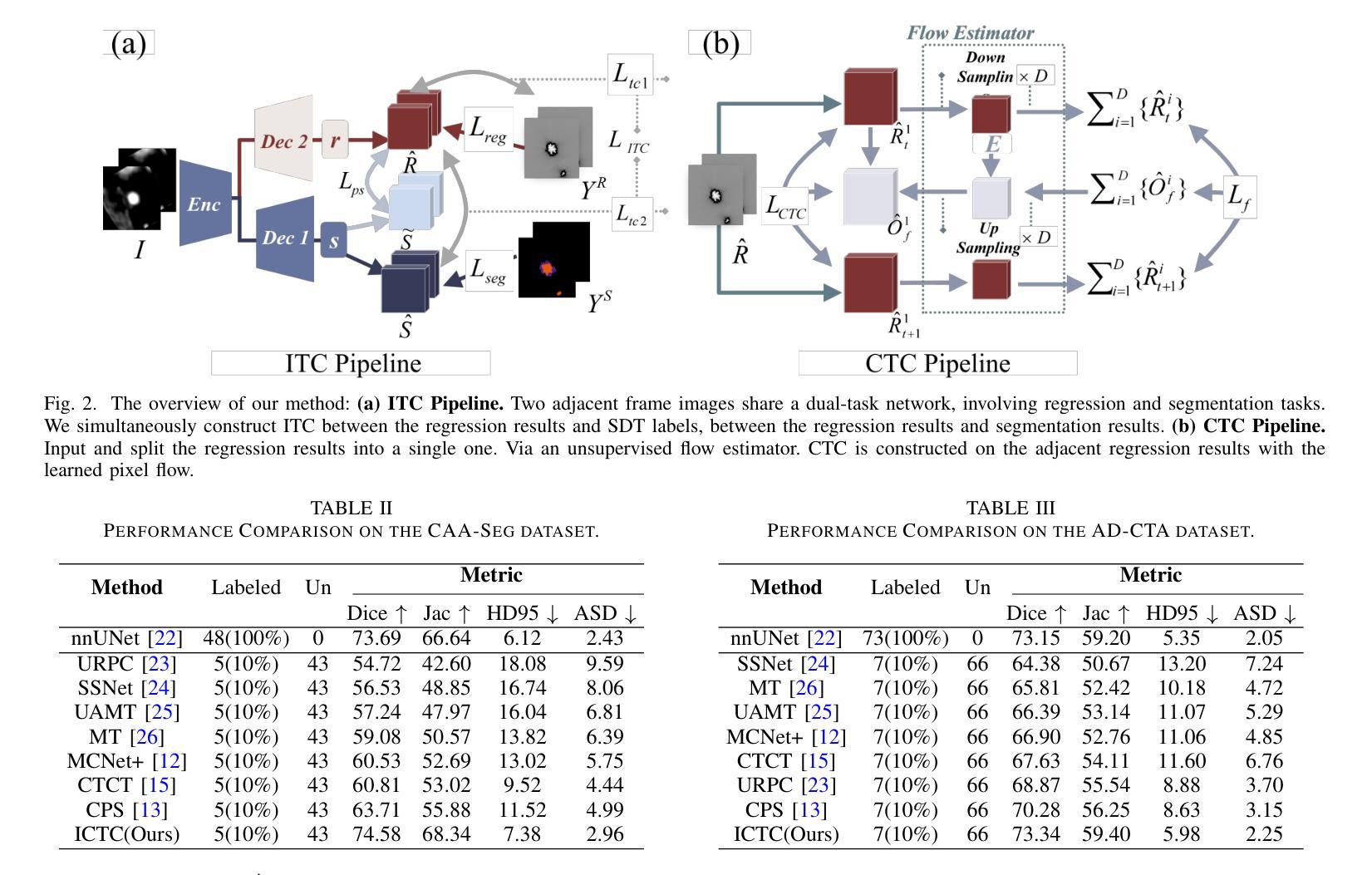
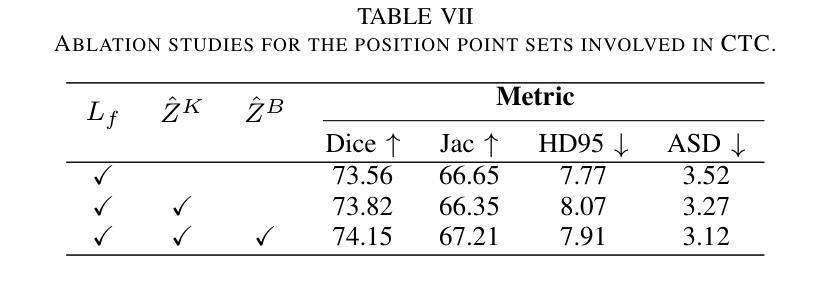
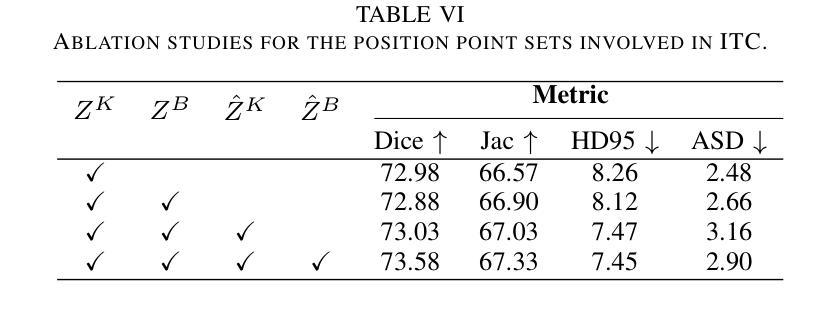
An Intra- and Cross-frame Topological Consistency Scheme for Semi-supervised Atherosclerotic Coronary Plaque Segmentation
Authors:Ziheng Zhang, Zihan Li, Dandan Shan, Yuehui Qiu, Qingqi Hong, Qingqiang Wu
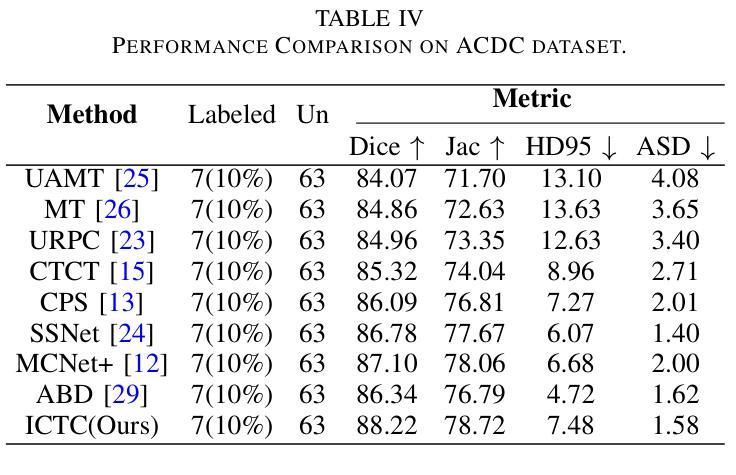
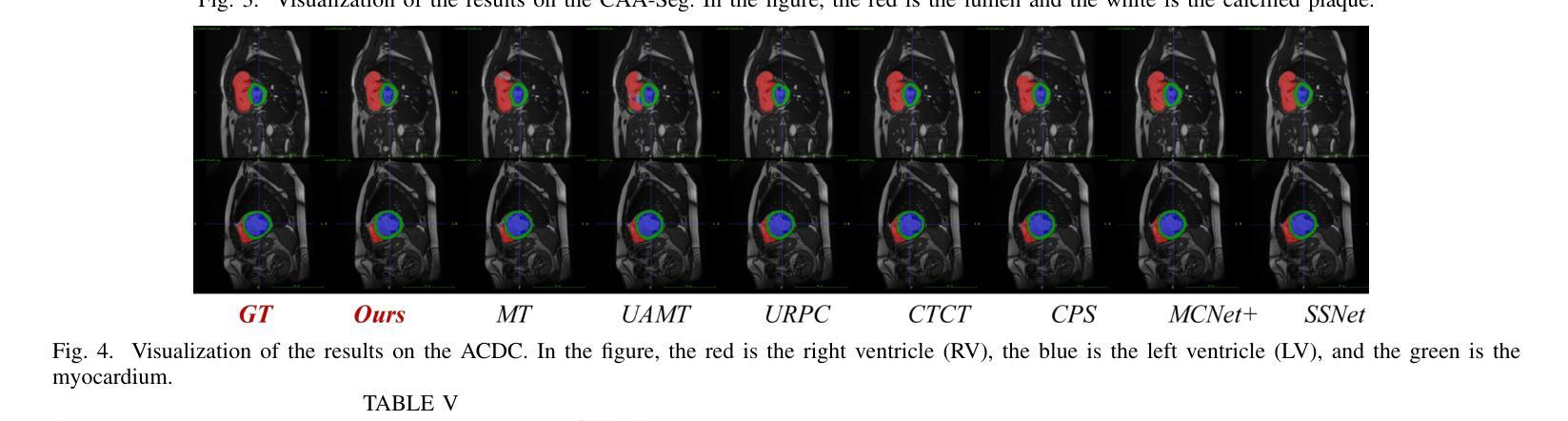
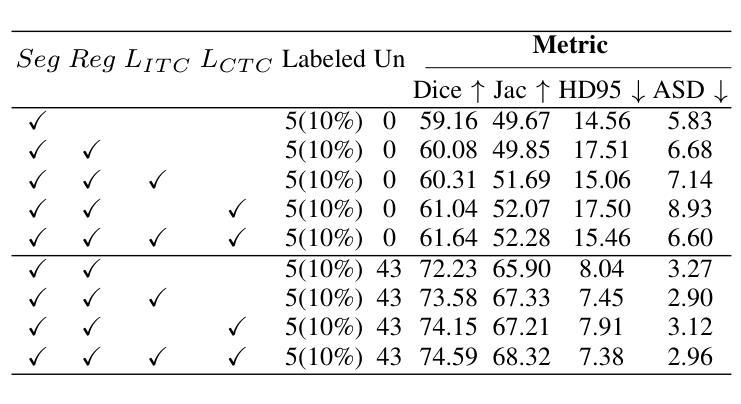
Enhancing the precision of segmenting coronary atherosclerotic plaques from CT Angiography (CTA) images is pivotal for advanced Coronary Atherosclerosis Analysis (CAA), which distinctively relies on the analysis of vessel cross-section images reconstructed via Curved Planar Reformation. This task presents significant challenges due to the indistinct boundaries and structures of plaques and blood vessels, leading to the inadequate performance of current deep learning models, compounded by the inherent difficulty in annotating such complex data. To address these issues, we propose a novel dual-consistency semi-supervised framework that integrates Intra-frame Topological Consistency (ITC) and Cross-frame Topological Consistency (CTC) to leverage labeled and unlabeled data. ITC employs a dual-task network for simultaneous segmentation mask and Skeleton-aware Distance Transform (SDT) prediction, achieving similar prediction of topology structure through consistency constraint without additional annotations. Meanwhile, CTC utilizes an unsupervised estimator for analyzing pixel flow between skeletons and boundaries of adjacent frames, ensuring spatial continuity. Experiments on two CTA datasets show that our method surpasses existing semi-supervised methods and approaches the performance of supervised methods on CAA. In addition, our method also performs better than other methods on the ACDC dataset, demonstrating its generalization.
提高冠状动脉粥样硬化斑块在CT血管造影(CTA)图像中的分割精度对于先进的冠状动脉粥样硬化分析(CAA)至关重要。CAA主要依赖于通过曲面平面重构技术重建的血管横截面图像的分析。这一任务面临重大挑战,因为斑块的边界和结构模糊不清,导致当前深度学习模型的性能不足,加上标注此类复杂数据的固有困难。为了解决这个问题,我们提出了一种新型的双一致性半监督框架,该框架结合了帧内拓扑一致性(ITC)和跨帧拓扑一致性(CTC),以利用有标签和无标签的数据。ITC采用双任务网络同时进行分割掩膜和骨架感知距离变换(SDT)预测,通过一致性约束实现类似的拓扑结构预测,无需额外的标注。同时,CTC利用无监督估计器分析相邻帧之间骨架和边界的像素流,确保空间连续性。在两个CTA数据集上的实验表明,我们的方法超越了现有的半监督方法,并接近了CAA方面的监督方法性能。此外,我们的方法在ACDC数据集上的表现也优于其他方法,证明了其泛化能力。
论文及项目相关链接
PDF Accepted by ICASSP 2025
Summary
该文本介绍了冠状动脉粥样硬化分析(CAA)中从CT血管造影(CTA)图像精确分割冠状动脉粥样硬化斑块的重要性。针对当前深度学习模型在标注复杂数据方面的不足,提出了一种新的双一致性半监督框架,该框架结合了帧内拓扑一致性(ITC)和跨帧拓扑一致性(CTC),以利用有标签和无标签的数据。实验表明,该方法在CTA数据集上的表现优于现有的半监督方法,并接近有监督方法在CAA上的表现。此外,该方法在ACDC数据集上的表现也优于其他方法,显示出其泛化能力。
Key Takeaways
- 精确分割冠状动脉粥样硬化斑块对冠状动脉粥样硬化分析(CAA)至关重要。
- 当前深度学习模型在标注复杂数据方面面临挑战。
- 提出了一种新的双一致性半监督框架,结合了ITC和CTC以提高性能。
- ITC通过双任务网络进行同时分割掩膜和骨架感知距离变换预测,实现拓扑结构的相似性预测。
- CTC利用无监督估计器分析相邻帧之间像素流与骨架和边界的关系,确保空间连续性。
- 在CTA数据集上的实验表明,该方法优于现有半监督方法,并接近有监督方法的性能。
点此查看论文截图

A Low-cost and Ultra-lightweight Binary Neural Network for Traffic Signal Recognition
Authors:Mingke Xiao, Yue Su, Liang Yu, Guanglong Qu, Yutong Jia, Yukuan Chang, Xu Zhang
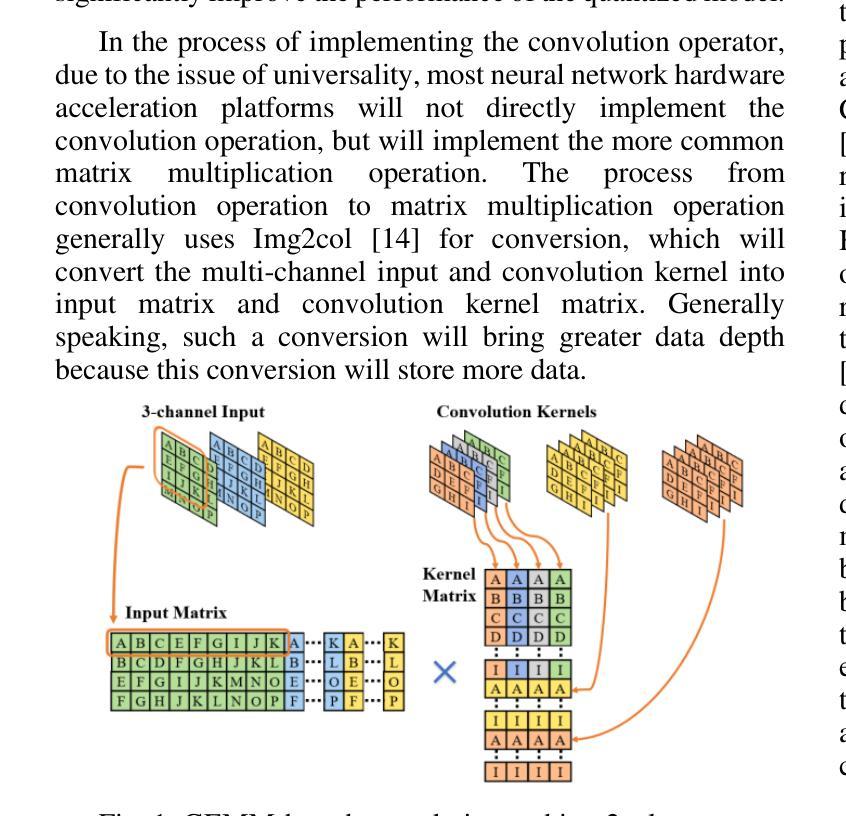
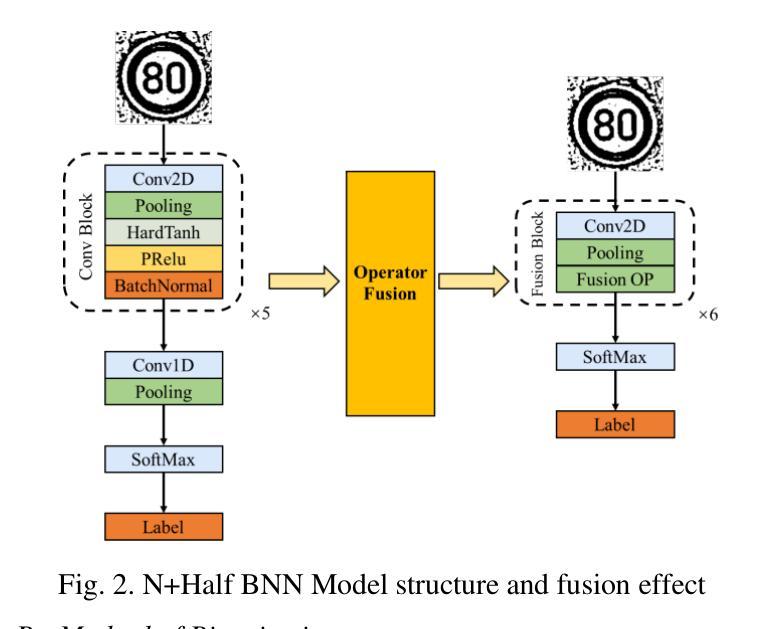
The deployment of neural networks in vehicle platforms and wearable Artificial Intelligence-of-Things (AIOT) scenarios has become a research area that has attracted much attention. With the continuous evolution of deep learning technology, many image classification models are committed to improving recognition accuracy, but this is often accompanied by problems such as large model resource usage, complex structure, and high power consumption, which makes it challenging to deploy on resource-constrained platforms. Herein, we propose an ultra-lightweight binary neural network (BNN) model designed for hardware deployment, and conduct image classification research based on the German Traffic Sign Recognition Benchmark (GTSRB) dataset. In addition, we also verify it on the Chinese Traffic Sign (CTS) and Belgian Traffic Sign (BTS) datasets. The proposed model shows excellent recognition performance with an accuracy of up to 97.64%, making it one of the best performing BNN models in the GTSRB dataset. Compared with the full-precision model, the accuracy loss is controlled within 1%, and the parameter storage overhead of the model is only 10% of that of the full-precision model. More importantly, our network model only relies on logical operations and low-bit width fixed-point addition and subtraction operations during the inference phase, which greatly simplifies the design complexity of the processing element (PE). Our research shows the great potential of BNN in the hardware deployment of computer vision models, especially in the field of computer vision tasks related to autonomous driving.
神经网络在车辆平台和可穿戴人工智能物联网(AIOT)场景的应用已成为一个备受关注的研究领域。随着深度学习技术的不断发展,许多图像分类模型致力于提高识别精度,但这常常伴随着模型资源使用量大、结构复杂、功耗高等问题,使得在资源受限的平台上的部署具有挑战性。针对这一问题,我们提出了一种专为硬件部署设计的超轻量级二进制神经网络(BNN)模型,并基于德国交通标志识别基准测试(GTSRB)数据集进行图像分类研究。此外,我们还对中国交通标志(CTS)和比利时交通标志(BTS)数据集进行了验证。该模型表现出优异的识别性能,准确率高达97.64%,成为GTSRB数据集中性能最佳的BNN模型之一。与全精度模型相比,精度损失控制在1%以内,模型参数存储开销仅为全精度模型的10%。更重要的是,我们的网络模型在推理阶段仅依赖逻辑操作和低位宽固定点加减运算,这极大地简化了处理元件(PE)的设计复杂度。我们的研究显示了BNN在计算机视觉模型的硬件部署中的巨大潜力,特别是在与自动驾驶相关的计算机视觉任务领域。
论文及项目相关链接
Summary
针对车辆平台和可穿戴人工智能物联网场景,提出一种超轻量级二进制神经网络模型进行图像分类研究,基于德国交通标志识别基准数据集进行验证,表现出卓越的性能和高达97.64%的识别准确率。与全精度模型相比,参数存储开销仅占其十分之一,推理阶段仅依赖逻辑操作和低位定点加减运算,大幅简化处理元件设计复杂度,展示二进制神经网络在自动驾驶相关计算机视觉任务的硬件部署中的潜力。
Key Takeaways
- 神经网络在车辆平台和可穿戴AIoT场景中的应用已成为研究热点。
- 深度学习技术在图像分类模型中的不断进步同时也带来了模型资源消耗大、结构复杂、功耗高等问题。
- 提出一种超轻量级二进制神经网络(BNN)模型,适用于硬件部署。
- 在德国交通标志识别基准数据集上验证,识别准确率高达97.64%,性能卓越。
- 与全精度模型相比,参数存储开销大幅降低,仅为其十分之一。
- 推理阶段仅依赖逻辑操作和低位定点加减运算,简化处理元件设计复杂度。
点此查看论文截图

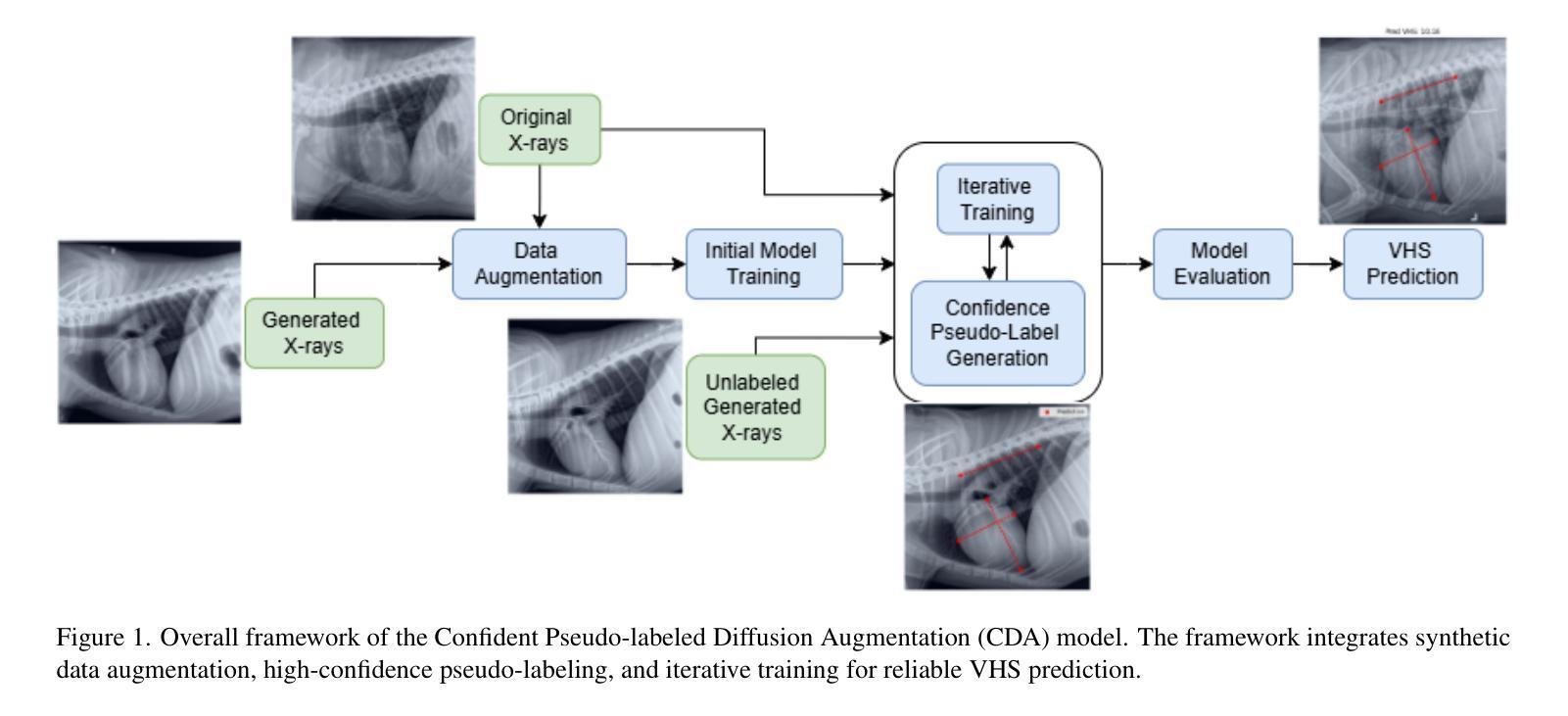
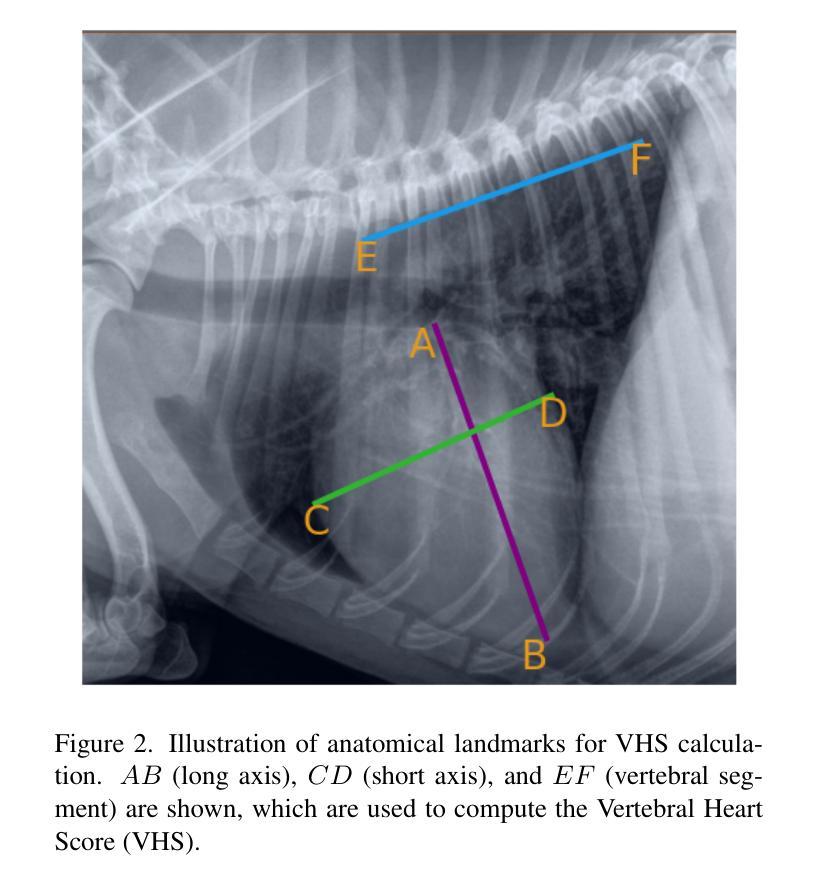
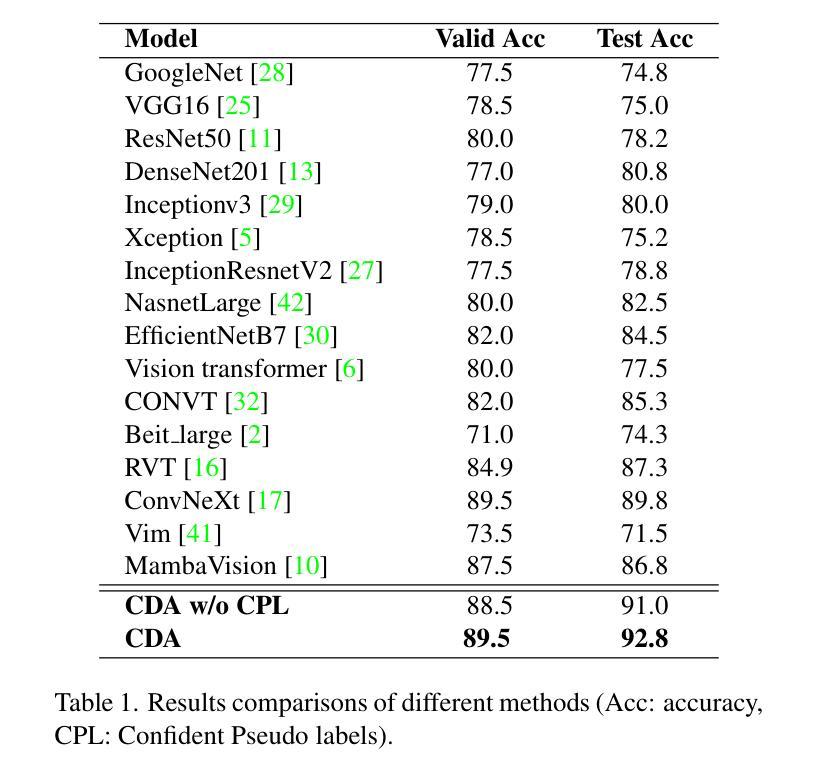
Confident Pseudo-labeled Diffusion Augmentation for Canine Cardiomegaly Detection
Authors:Shiman Zhang, Lakshmikar Reddy Polamreddy, Youshan Zhang
Canine cardiomegaly, marked by an enlarged heart, poses serious health risks if undetected, requiring accurate diagnostic methods. Current detection models often rely on small, poorly annotated datasets and struggle to generalize across diverse imaging conditions, limiting their real-world applicability. To address these issues, we propose a Confident Pseudo-labeled Diffusion Augmentation (CDA) model for identifying canine cardiomegaly. Our approach addresses the challenge of limited high-quality training data by employing diffusion models to generate synthetic X-ray images and annotate Vertebral Heart Score key points, thereby expanding the dataset. We also employ a pseudo-labeling strategy with Monte Carlo Dropout to select high-confidence labels, refine the synthetic dataset, and improve accuracy. Iteratively incorporating these labels enhances the model’s performance, overcoming the limitations of existing approaches. Experimental results show that the CDA model outperforms traditional methods, achieving state-of-the-art accuracy in canine cardiomegaly detection. The code implementation is available at https://github.com/Shira7z/CDA.
犬类心脏肥大表现为心脏增大,如果未被检测出来,会带来严重的健康风险,因此需要准确的诊断方法。当前的检测模型通常依赖于小且标注不佳的数据集,难以在不同成像条件下进行推广,限制了它们在现实世界中的应用。为了解决这些问题,我们提出了一种用于识别犬类心脏肥大的自信伪标记扩散增强(CDA)模型。我们的方法通过采用扩散模型生成合成X射线图像并标注椎体心脏评分关键点,从而解决高质量训练数据有限的问题,以此扩大数据集。我们还采用了一种带有蒙特卡洛Dropout的伪标记策略,选择高置信度标签,对合成数据集进行精炼,提高了准确性。迭代地融入这些标签提高了模型的性能,克服了现有方法的局限性。实验结果表明,CDA模型优于传统方法,在犬类心脏肥大检测方面达到了最先进的准确度。代码实现可访问https://github.com/Shira7z/CDA。
论文及项目相关链接
PDF WACV workshop
Summary
当前犬类心脏病检测模型受限于小型且标注不全的数据集,难以适应不同成像条件,实际应用受限。为解决这些问题,我们提出一种名为Confident Pseudo-labeled Diffusion Augmentation(CDA)的检测模型。该模型通过扩散模型生成合成X射线图像并标注心脏分数关键点,以扩充数据集并提升准确性。实验结果证明,CDA模型表现优于传统方法,达到犬类心脏病检测的最先进水平。代码实现可访问:链接。
Key Takeaways
- 犬心脏病若未检测可能带来健康风险。
- 当前检测模型受限于小型数据集和难以适应不同成像条件的问题。
- CDA模型通过扩散模型生成合成X射线图像并标注心脏分数关键点以扩充数据集。
- CDA模型采用伪标签策略与Monte Carlo Dropout技术提升数据质量及模型准确性。
- CDA模型性能优于传统方法,达到犬心脏病检测的最先进水平。
点此查看论文截图
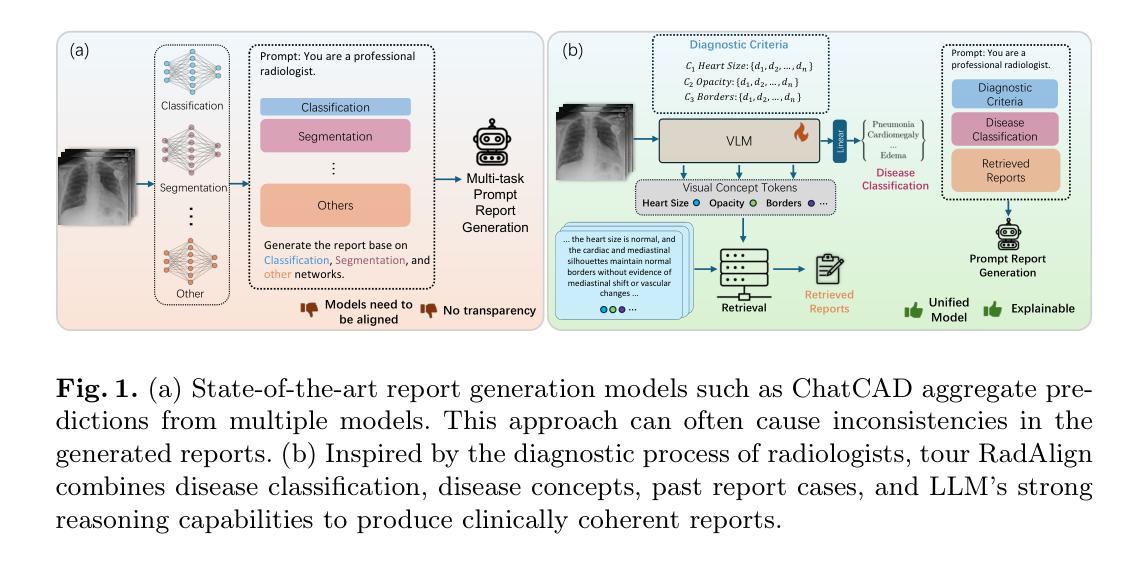
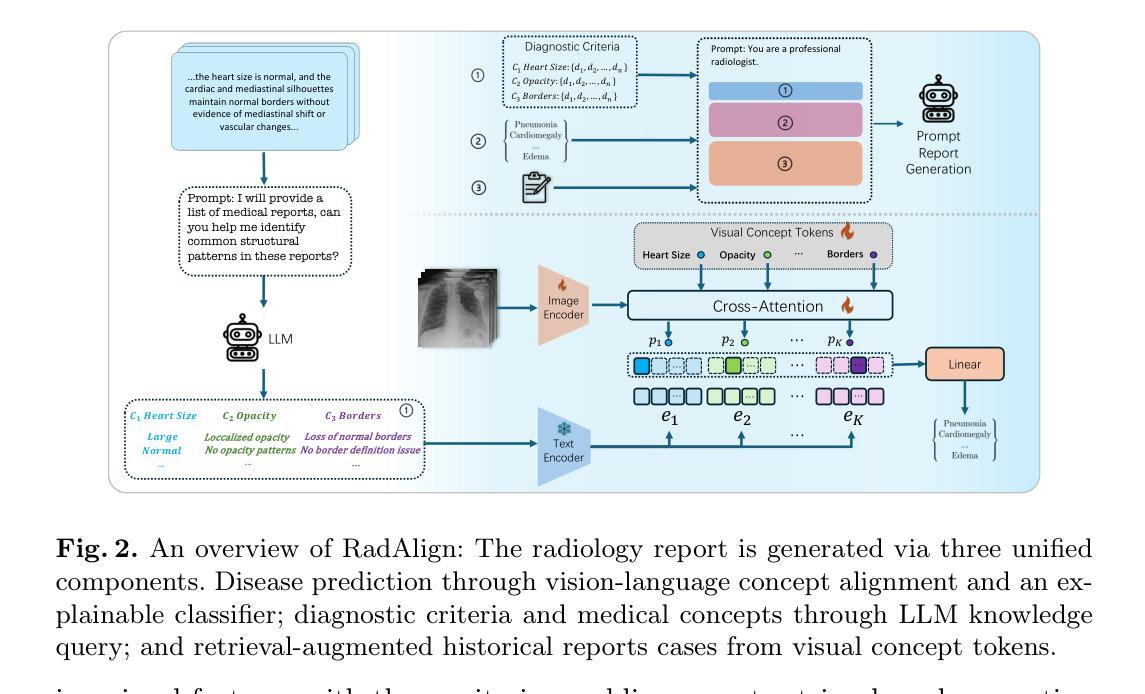
RadAlign: Advancing Radiology Report Generation with Vision-Language Concept Alignment
Authors:Difei Gu, Yunhe Gao, Yang Zhou, Mu Zhou, Dimitris Metaxas
Automated chest radiographs interpretation requires both accurate disease classification and detailed radiology report generation, presenting a significant challenge in the clinical workflow. Current approaches either focus on classification accuracy at the expense of interpretability or generate detailed but potentially unreliable reports through image captioning techniques. In this study, we present RadAlign, a novel framework that combines the predictive accuracy of vision-language models (VLMs) with the reasoning capabilities of large language models (LLMs). Inspired by the radiologist’s workflow, RadAlign first employs a specialized VLM to align visual features with key medical concepts, achieving superior disease classification with an average AUC of 0.885 across multiple diseases. These recognized medical conditions, represented as text-based concepts in the aligned visual-language space, are then used to prompt LLM-based report generation. Enhanced by a retrieval-augmented generation mechanism that grounds outputs in similar historical cases, RadAlign delivers superior report quality with a GREEN score of 0.678, outperforming state-of-the-art methods’ 0.634. Our framework maintains strong clinical interpretability while reducing hallucinations, advancing automated medical imaging and report analysis through integrated predictive and generative AI. Code is available at https://github.com/difeigu/RadAlign.
自动化胸部放射影像解读既需要精确的疾病分类,也需要生成详细的放射学报告,这在临床工作流程中面临重大挑战。当前的方法要么侧重于分类准确性而牺牲了可解释性,要么通过图像描述技术生成详细但可能不可靠的报告。在本研究中,我们提出了RadAlign,这是一种结合视觉语言模型(VLM)的预测准确性以及大型语言模型(LLM)的推理能力的新型框架。RadAlign受放射科医生工作流程的启发,首先采用专用的VLM对齐视觉特征与关键医学概念,实现对多种疾病的卓越疾病分类,平均AUC(曲线下面积)为0.885。这些被识别的医学状况,以对齐的视觉语言空间中的基于文本的概念表示,然后用于提示基于LLM的报告生成。通过增强以相似历史病例为基础的检索增强生成机制,RadAlign在报告质量上表现出卓越的性能,GREEN得分为0.678,超过了最新方法(0.634)。我们的框架在保持强大的临床可解释性的同时,减少了虚构情况,通过集成预测和生成人工智能,推动了自动医疗成像和报告分析的发展。代码可在链接中找到。
论文及项目相关链接
Summary
本文介绍了一种名为RadAlign的新型框架,该框架结合了视觉语言模型(VLMs)的预测精度和大语言模型(LLMs)的推理能力,用于自动解读胸部X光片。RadAlign不仅能准确分类疾病,还能生成详细的放射学报告。通过专门的VLM对齐视觉特征与关键医学概念,实现多疾病平均AUC值为0.885的优越疾病分类。然后,使用LLM生成报告,输出基于类似历史病例的输出,提高报告质量。RadAlign框架既具有强大的临床可解释性,又减少了虚构现象,通过预测和生成式人工智能的集成,推动了自动医学成像和报告分析的发展。
Key Takeaways
- RadAlign框架结合了视觉语言模型(VLMs)和大语言模型(LLMs)的优势,用于自动解读胸部X光片。
- RadAlign实现高准确性的疾病分类,平均AUC值为0.885。
- 该框架能够生成详细的放射学报告,报告质量高于现有方法。
- RadAlign通过专门设计的VLM对齐视觉特征与医学概念,提高了疾病分类的准确性。
- LLM用于生成报告,可以输出基于类似历史病例的信息,进一步提高报告质量。
- RadAlign框架具有强大的临床可解释性,减少了虚构现象。
点此查看论文截图

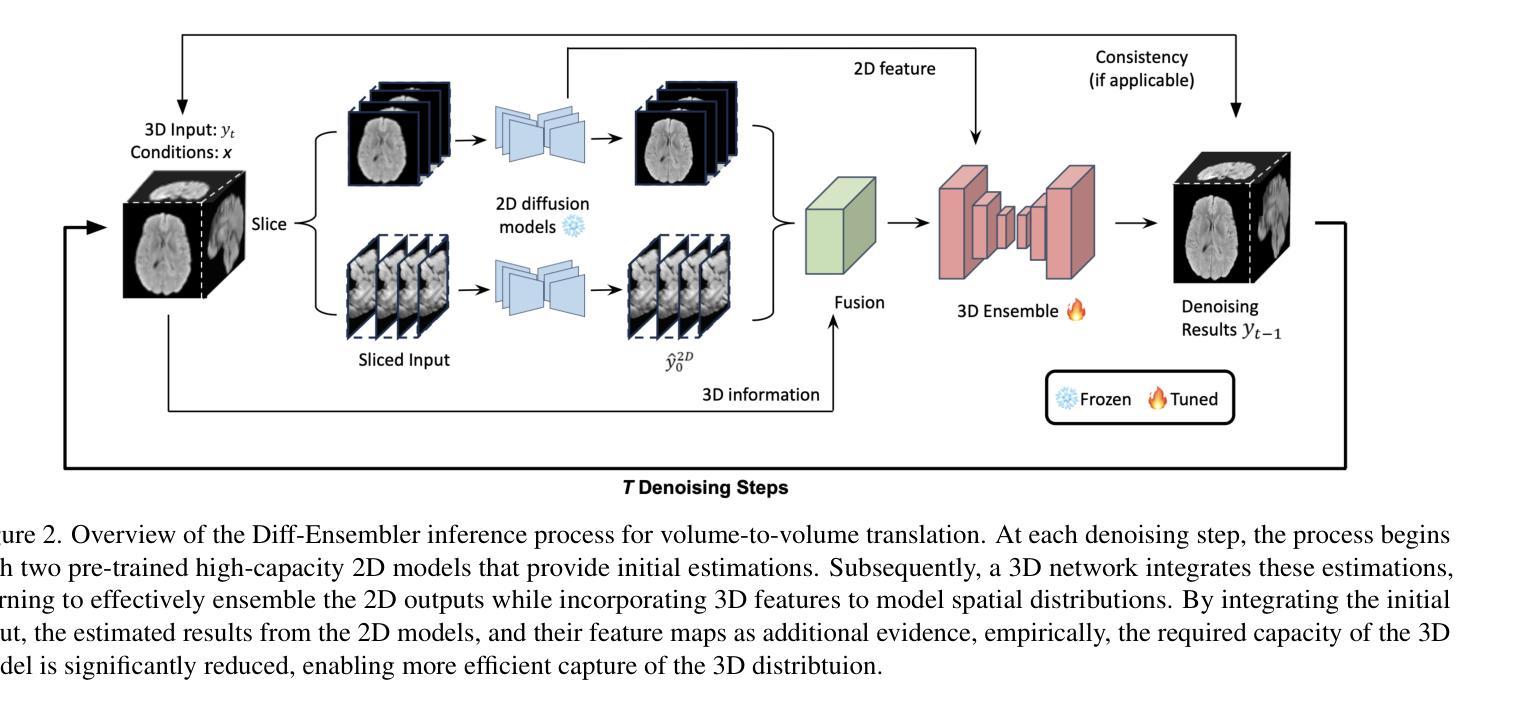
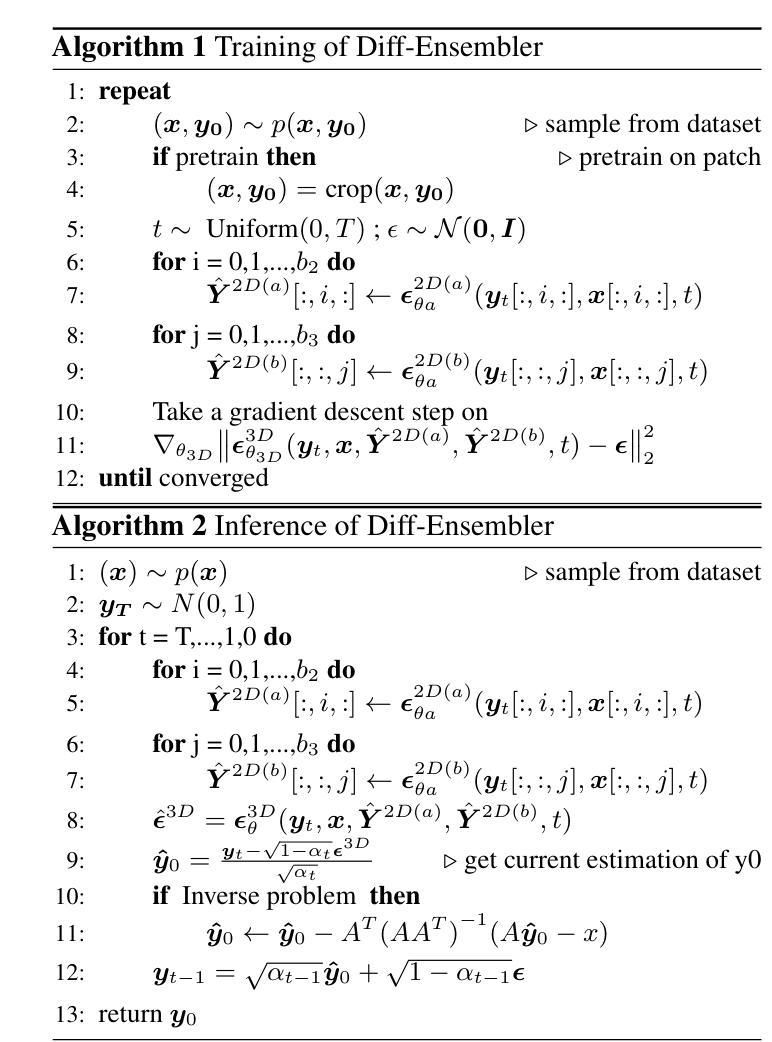
Diff-Ensembler: Learning to Ensemble 2D Diffusion Models for Volume-to-Volume Medical Image Translation
Authors:Xiyue Zhu, Dou Hoon Kwark, Ruike Zhu, Kaiwen Hong, Yiqi Tao, Shirui Luo, Yudu Li, Zhi-Pei Liang, Volodymyr Kindratenko
Despite success in volume-to-volume translations in medical images, most existing models struggle to effectively capture the inherent volumetric distribution using 3D representations. The current state-of-the-art approach combines multiple 2D-based networks through weighted averaging, thereby neglecting the 3D spatial structures. Directly training 3D models in medical imaging presents significant challenges due to high computational demands and the need for large-scale datasets. To address these challenges, we introduce Diff-Ensembler, a novel hybrid 2D-3D model for efficient and effective volumetric translations by ensembling perpendicularly trained 2D diffusion models with a 3D network in each diffusion step. Moreover, our model can naturally be used to ensemble diffusion models conditioned on different modalities, allowing flexible and accurate fusion of input conditions. Extensive experiments demonstrate that Diff-Ensembler attains superior accuracy and volumetric realism in 3D medical image super-resolution and modality translation. We further demonstrate the strength of our model’s volumetric realism using tumor segmentation as a downstream task.
尽管在医学图像的体积到体积转换中取得了一定的成功,但大多数现有模型在利用3D表示捕捉内在的体积分布时仍面临困难。当前最先进的方法是通过加权平均组合多个基于2D的网络,从而忽略了3D空间结构。直接在医学成像中训练3D模型面临着巨大的计算需求和大规模数据集的需求,这带来了很大的挑战。为了应对这些挑战,我们引入了Diff-Ensembler,这是一种新型的混合2D-3D模型,它通过在每个扩散步骤中将垂直训练的2D扩散模型与3D网络相结合,实现了高效且有效的体积转换。此外,我们的模型可以自然地用于组合不同模态的扩散模型,实现输入条件的灵活和准确融合。大量实验表明,Diff-Ensembler在3D医学图像超分辨率和模态转换方面达到了较高的准确性和体积真实性。我们进一步通过肿瘤分割作为下游任务来展示我们的模型在体积真实性方面的优势。
论文及项目相关链接
Summary
本文提出一种新型混合2D-3D模型——Diff-Ensembler,旨在通过集成垂直训练的2D扩散模型与3D网络,实现高效且有效的体积转换。该模型能够融合不同模态的扩散模型,具有灵活且精确的输入条件融合能力。实验证明,Diff-Ensembler在3D医学图像超分辨率和模态转换方面达到优越的准确性及体积现实性。
Key Takeaways
- 现有模型在医学图像体积转换时难以有效捕捉内在体积分布,缺乏3D表示。
- 当前最新方法是通过加权平均多个基于2D的网络,忽略了3D空间结构。
- 直接训练3D医学成像模型面临高计算需求和大规模数据集挑战。
- Diff-Ensembler是一种新型混合2D-3D模型,通过集成垂直训练的2D扩散模型和3D网络,实现高效和有效的体积转换。
- Diff-Ensembler能自然融合不同模态的扩散模型,实现灵活且精确的输入条件融合。
- 实验证明Diff-Ensembler在3D医学图像超分辨率和模态转换方面具有优越性能和体积现实性。
点此查看论文截图

FedSemiDG: Domain Generalized Federated Semi-supervised Medical Image Segmentation
Authors:Zhipeng Deng, Zhe Xu, Tsuyoshi Isshiki, Yefeng Zheng
Medical image segmentation is challenging due to the diversity of medical images and the lack of labeled data, which motivates recent developments in federated semi-supervised learning (FSSL) to leverage a large amount of unlabeled data from multiple centers for model training without sharing raw data. However, what remains under-explored in FSSL is the domain shift problem which may cause suboptimal model aggregation and low effectivity of the utilization of unlabeled data, eventually leading to unsatisfactory performance in unseen domains. In this paper, we explore this previously ignored scenario, namely domain generalized federated semi-supervised learning (FedSemiDG), which aims to learn a model in a distributed manner from multiple domains with limited labeled data and abundant unlabeled data such that the model can generalize well to unseen domains. We present a novel framework, Federated Generalization-Aware SemiSupervised Learning (FGASL), to address the challenges in FedSemiDG by effectively tackling critical issues at both global and local levels. Globally, we introduce Generalization-Aware Aggregation (GAA), assigning adaptive weights to local models based on their generalization performance. Locally, we use a Dual-Teacher Adaptive Pseudo Label Refinement (DR) strategy to combine global and domain-specific knowledge, generating more reliable pseudo labels. Additionally, Perturbation-Invariant Alignment (PIA) enforces feature consistency under perturbations, promoting domain-invariant learning. Extensive experiments on three medical segmentation tasks (cardiac MRI, spine MRI and bladder cancer MRI) demonstrate that our method significantly outperforms state-of-the-art FSSL and domain generalization approaches, achieving robust generalization on unseen domains.
医学图像分割面临诸多挑战,主要是由于医学图像的多样性和缺乏标记数据。这推动了联邦半监督学习(FSSL)的近期发展,以利用多个中心的大量未标记数据进行模型训练,而无需共享原始数据。然而,在FSSL中仍未得到充分探索的是域偏移问题,它可能导致模型聚合不佳以及利用未标记数据的效果低下,最终导致在未见过的领域中的性能不佳。本文探索了以前被忽略的场景,即领域泛化联邦半监督学习(FedSemiDG),其目标是从多个领域以分布式方式学习模型,利用有限的标记数据和丰富的未标记数据,使模型能够很好地泛化到未见过的领域。我们提出了一种新的框架——联邦泛化感知半监督学习(FGASL),以应对FedSemiDG中的挑战,在全局和局部层面有效解决关键问题。在全局层面,我们引入了泛化感知聚合(GAA),根据局部模型的泛化性能分配自适应权重。在局部层面,我们使用双教师自适应伪标签细化(DR)策略来结合全局和领域特定知识,生成更可靠的伪标签。此外,扰动不变对齐(PIA)强制执行扰动下的特征一致性,促进领域不变学习。在心脏MRI、脊柱MRI和膀胱癌MRI等三个医学分割任务上的大量实验表明,我们的方法显著优于最新的FSSL和领域泛化方法,在未见过的领域上实现了稳健的泛化。
论文及项目相关链接
PDF 17 pages
摘要
医学图像分割面临多样性图像与缺乏标注数据的挑战。联邦半监督学习(FSSL)的发展旨在利用多个中心的大量未标注数据进行模型训练,但不分享原始数据。然而,FSSL中尚未探索的问题是领域偏移,可能导致模型聚合不佳、利用未标注数据效率低,以及在未见领域表现不佳。本文探索了之前被忽略的场景——领域广义联邦半监督学习(FedSemiDG),旨在从多个领域以分布式方式学习模型,使用有限的标注数据和丰富的未标注数据,使模型能在未见领域表现良好。我们提出了一个新的框架——联邦泛化感知半监督学习(FGASL),通过全球和本地层面有效应对FedSemiDG中的挑战。全球层面,我们引入泛化感知聚合(GAA),根据泛化性能为本地模型分配自适应权重。本地层面,我们使用双教师自适应伪标签细化(DR)策略,结合全局和领域特定知识,生成更可靠的伪标签。此外,扰动不变对齐(PIA)在扰动下强制特征一致性,促进领域不变学习。在三个医学分割任务(心脏MRI、脊柱MRI和膀胱癌MRI)上的广泛实验表明,我们的方法显著优于最新的FSSL和领域泛化方法,在未见领域实现稳健的泛化。
关键见解
- 医学图像分割面临多样性图像和缺乏标注数据的挑战。
- 联邦半监督学习(FSSL)在利用多中心大量未标注数据进行模型训练方面展现出潜力,但领域偏移问题尚未得到充分探索。
- 提出了领域广义联邦半监督学习(FedSemiDG)的概念,旨在学习能适应未见领域的模型。
- 引入了一个新的框架——联邦泛化感知半监督学习(FGASL),结合全球和本地策略来应对FedSemiDG中的挑战。
- 在全球层面,通过泛化感知聚合(GAA)根据泛化性能分配自适应权重给本地模型。
- 在本地层面,采用双教师自适应伪标签细化(DR)策略,结合全局和领域特定知识,提高伪标签的可靠性。
- 扰动不变对齐(PIA)技术强制特征在扰动下保持一致性,促进领域不变学习,提高了模型的泛化能力。
点此查看论文截图

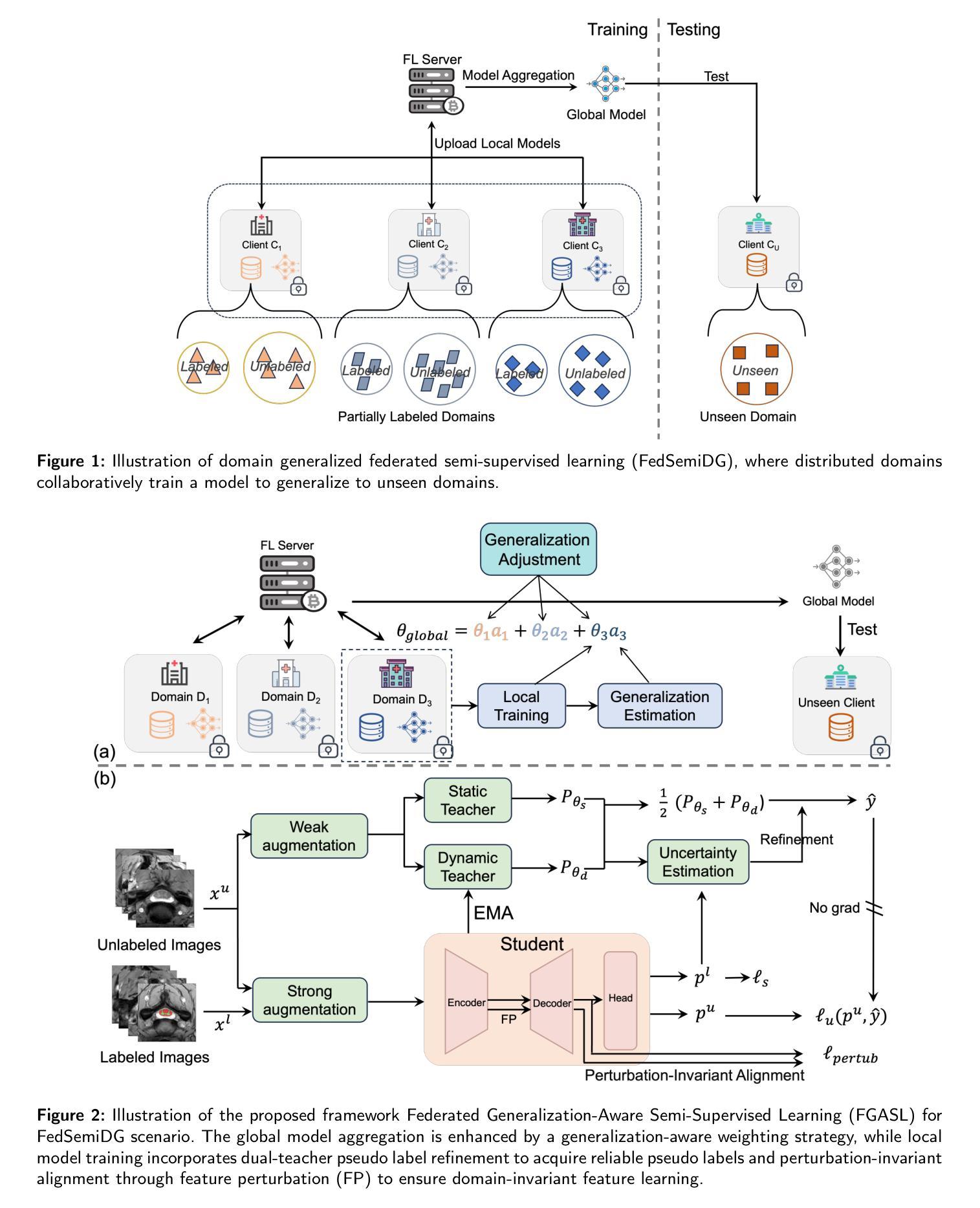
Bigger Isn’t Always Better: Towards a General Prior for Medical Image Reconstruction
Authors:Lukas Glaszner, Martin Zach
Diffusion model have been successfully applied to many inverse problems, including MRI and CT reconstruction. Researchers typically re-purpose models originally designed for unconditional sampling without modifications. Using two different posterior sampling algorithms, we show empirically that such large networks are not necessary. Our smallest model, effectively a ResNet, performs almost as good as an attention U-Net on in-distribution reconstruction, while being significantly more robust towards distribution shifts. Furthermore, we introduce models trained on natural images and demonstrate that they can be used in both MRI and CT reconstruction, out-performing model trained on medical images in out-of-distribution cases. As a result of our findings, we strongly caution against simply re-using very large networks and encourage researchers to adapt the model complexity to the respective task. Moreover, we argue that a key step towards a general diffusion-based prior is training on natural images.
扩散模型已成功应用于许多反问题,包括MRI和CT重建。研究者通常将原本为无条件采样设计的模型稍加改动后重新用于新的应用场景。通过使用两种不同的后采样算法,我们从实证角度证明,其实并不需要如此庞大的网络结构。我们最小的模型(实质上是一个ResNet)在分布内的重建上表现与注意力U-Net几乎一样好,同时对于分布转移表现出更强的稳健性。此外,我们引入了基于自然图像训练的模型,并证明其可用于MRI和CT重建,在分布外的案例中表现优于基于医学图像训练的模型。基于我们的发现,我们强烈反对简单地重复使用大型网络结构,并鼓励研究者根据各自任务调整模型复杂度。此外,我们认为面向通用扩散先验的关键步骤是在自然图像上进行训练。
论文及项目相关链接
PDF To appear in the German Conference on Pattern Recognition proceedings. Code available at https://github.com/VLOGroup/bigger-isnt-always-better
Summary
扩散模型已成功应用于MRI和CT重建等多个反问题。研究通常直接使用原本设计用于无条件采样的模型,而无需修改。通过两种不同的后采样算法,我们发现大型网络并非必需。我们的最小模型(即ResNet)在内部分布重建方面的表现几乎与注意力U-Net一样好,同时在分布转移方面表现出更强的稳健性。此外,我们展示了经自然图像训练模型在MRI和CT重建中的应用,并在外部分布情况下优于医学图像训练模型。因此,我们强烈建议不要简单地重复使用大型网络,并鼓励研究人员根据各自任务调整模型复杂度。此外,我们认为朝着基于扩散的先验的关键一步是在自然图像上进行训练。
Key Takeaways
- 扩散模型已成功应用于MRI和CT重建等反问题。
- 研究中通常直接使用无条件采样的模型,无需修改。
- 通过实验比较,发现大型网络在重建任务中并非必需。最小模型表现与更复杂的注意力U-Net相似或更佳。
- 对于内部分布重建任务,最小模型在性能上更为稳健。
- 通过自然图像训练的模型在MRI和CT重建中表现出良好的通用性,尤其在外部分布情况下性能更佳。
- 对使用大型网络的建议持谨慎态度,应适应性地调整模型复杂度以适应任务需求。
点此查看论文截图

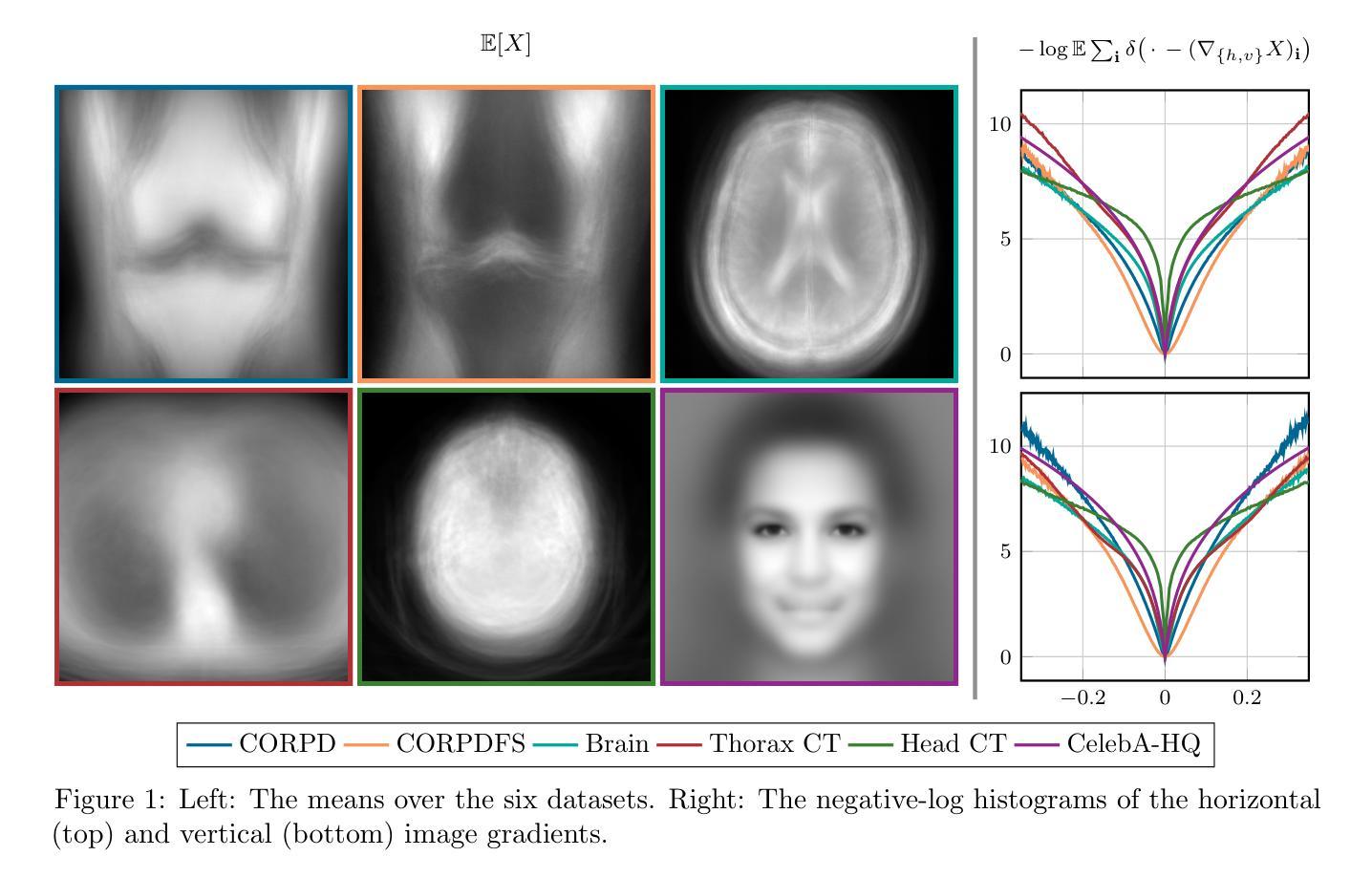
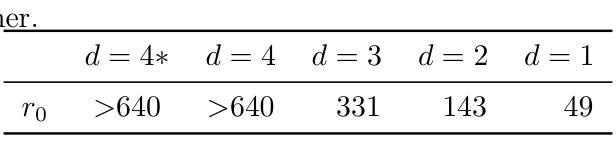
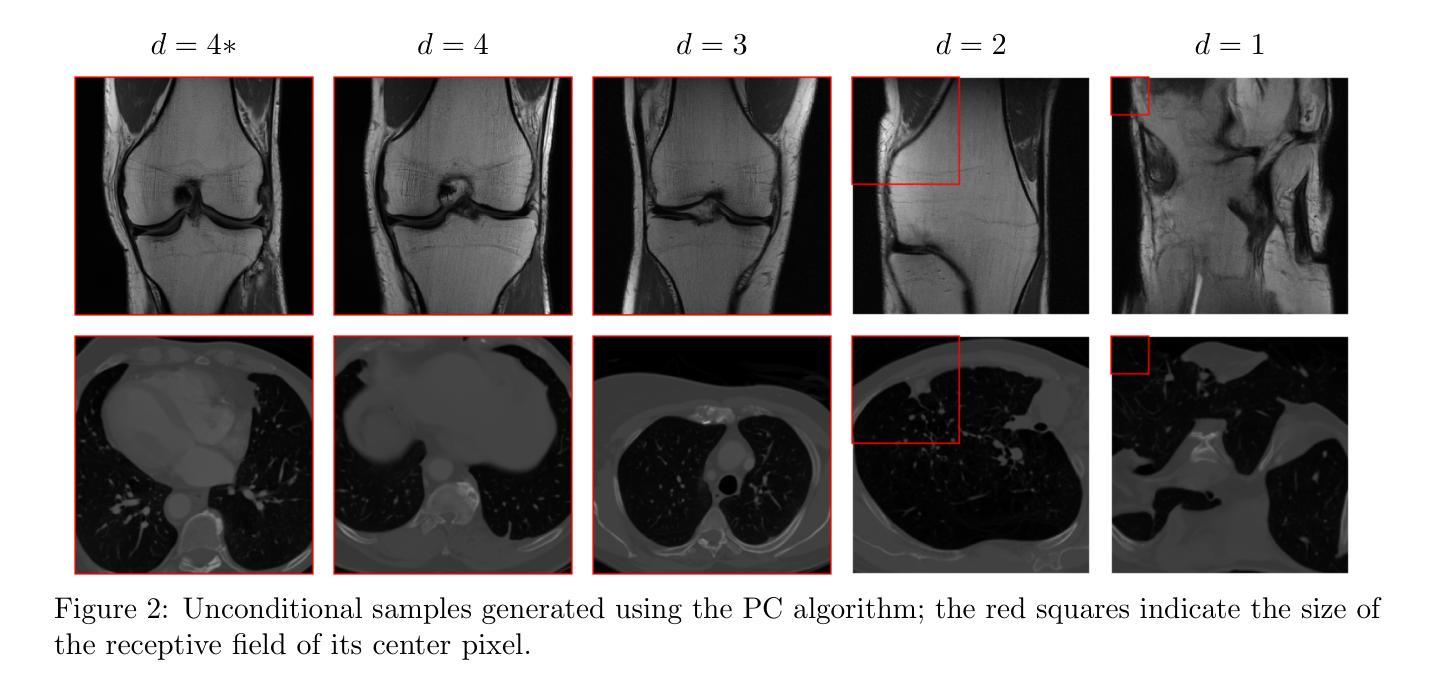
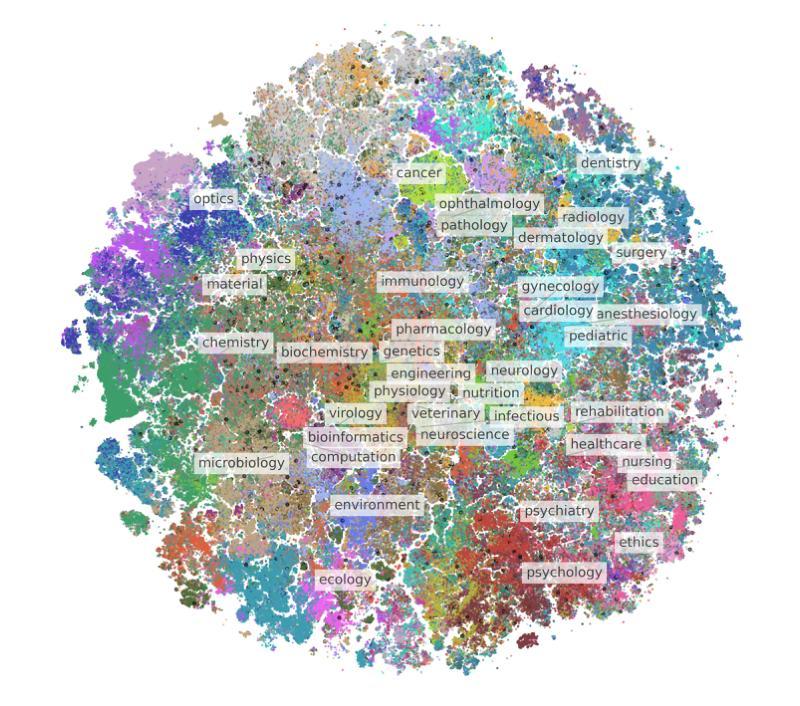
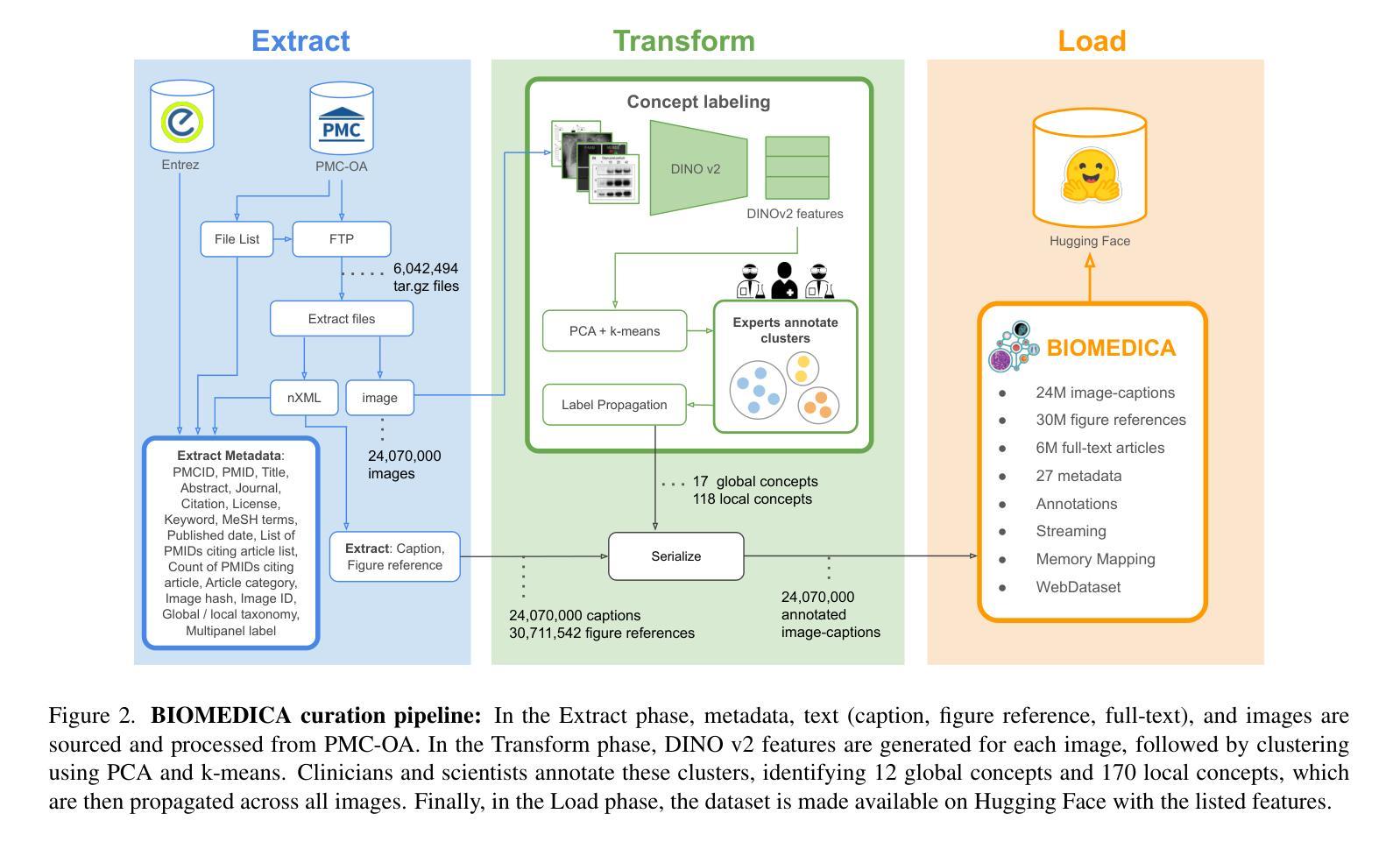
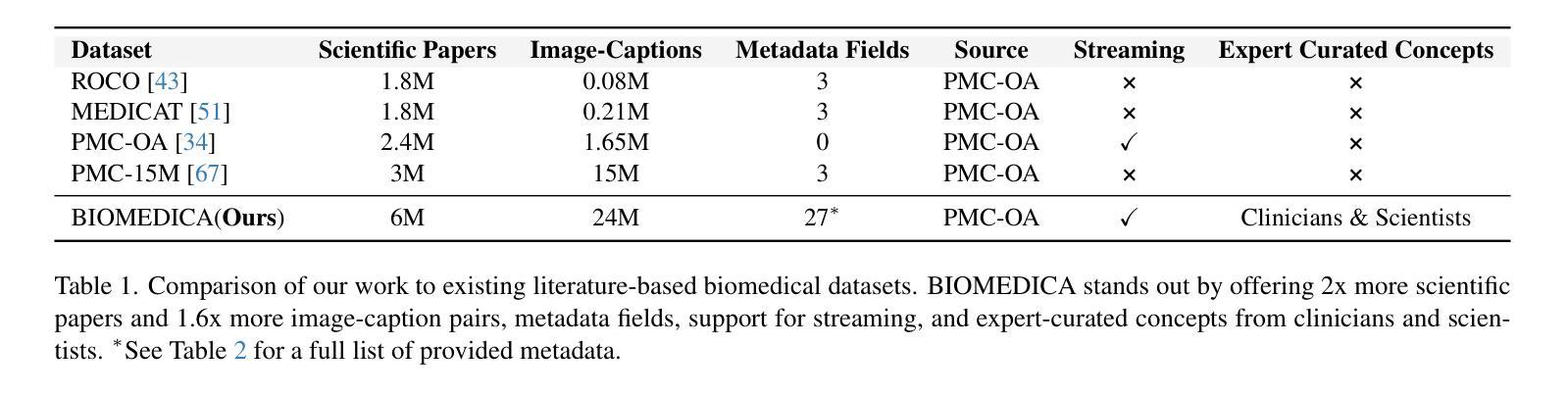
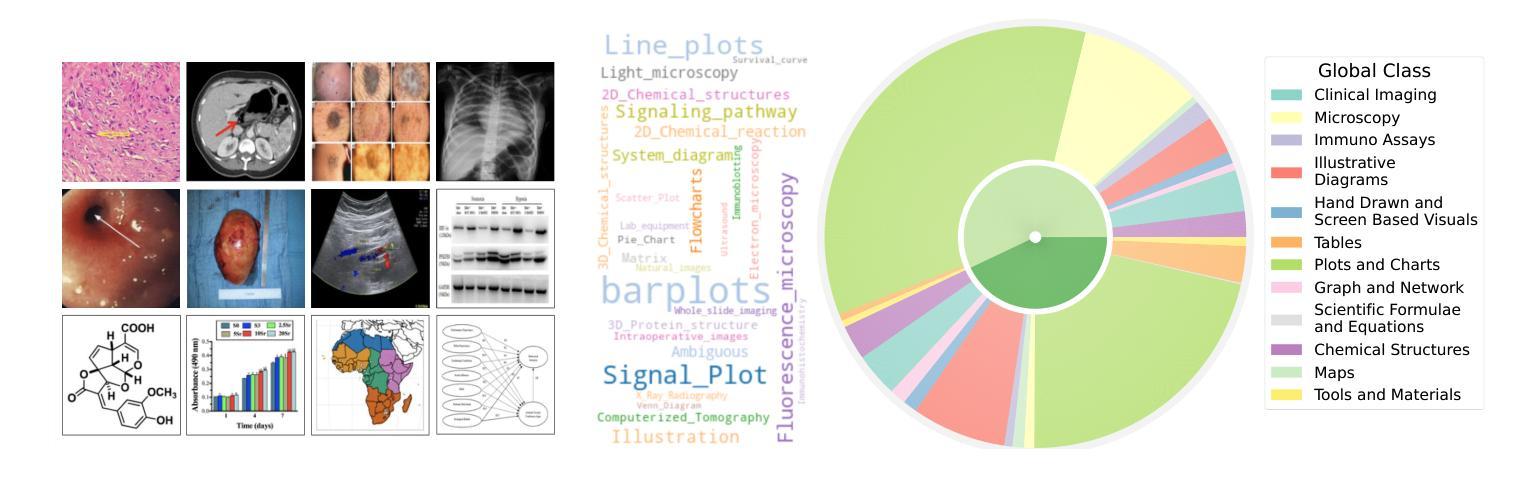
BIOMEDICA: An Open Biomedical Image-Caption Archive, Dataset, and Vision-Language Models Derived from Scientific Literature
Authors:Alejandro Lozano, Min Woo Sun, James Burgess, Liangyu Chen, Jeffrey J Nirschl, Jeffrey Gu, Ivan Lopez, Josiah Aklilu, Austin Wolfgang Katzer, Collin Chiu, Anita Rau, Xiaohan Wang, Yuhui Zhang, Alfred Seunghoon Song, Robert Tibshirani, Serena Yeung-Levy
The development of vision-language models (VLMs) is driven by large-scale and diverse multimodal datasets. However, progress toward generalist biomedical VLMs is limited by the lack of annotated, publicly accessible datasets across biology and medicine. Existing efforts are restricted to narrow domains, missing the full diversity of biomedical knowledge encoded in scientific literature. To address this gap, we introduce BIOMEDICA, a scalable, open-source framework to extract, annotate, and serialize the entirety of the PubMed Central Open Access subset into an easy-to-use, publicly accessible dataset. Our framework produces a comprehensive archive with over 24 million unique image-text pairs from over 6 million articles. Metadata and expert-guided annotations are also provided. We demonstrate the utility and accessibility of our resource by releasing BMCA-CLIP, a suite of CLIP-style models continuously pre-trained on the BIOMEDICA dataset via streaming, eliminating the need to download 27 TB of data locally. On average, our models achieve state-of-the-art performance across 40 tasks - spanning pathology, radiology, ophthalmology, dermatology, surgery, molecular biology, parasitology, and cell biology - excelling in zero-shot classification with a 6.56% average improvement (as high as 29.8% and 17.5% in dermatology and ophthalmology, respectively), and stronger image-text retrieval, all while using 10x less compute. To foster reproducibility and collaboration, we release our codebase and dataset for the broader research community.
视觉语言模型(VLMs)的发展是由大规模和多样化的多模态数据集驱动的。然而,通用生物医学VLMs的进展受限于生物学和医学领域缺乏公开访问的注释数据集。现有的努力仅限于狭窄的领域,错过了科学文献中编码的完整生物医学知识多样性。为了弥补这一差距,我们推出了BIOMEDICA,这是一个可扩展的开源框架,用于提取、注释和序列化PubMed Central Open Access子集的完整内容,使其成为易于使用、可公开访问的数据集。我们的框架产生了包含超过2.4亿个唯一图像文本对的综合档案,涵盖超过6百万篇文章。我们还提供了元数据和专业指导的注释。我们通过发布BMCA-CLIP来证明我们资源的实用性和可访问性,BMCA-CLIP是一套CLIP风格的模型,在BIOMEDICA数据集上通过流式传输进行连续预训练,无需下载本地存储的27TB数据。平均而言,我们的模型在涵盖病理学、放射学、眼科、皮肤科、外科、分子生物学、寄生虫学和细胞生物学等领域的四十多项任务上达到了领先水平,尤其在零样本分类方面取得了平均改进6.56%(皮肤病学和眼科分别高达29.8%和17.5%),并且图像文本检索能力更强,同时使用的计算资源减少了十倍。为了促进可复制性和合作,我们向更广泛的研究社区发布我们的代码和数据集。
论文及项目相关链接
Summary
BIOMEDICA框架解决了生物医学视觉语言模型(VLMs)发展的数据瓶颈问题。它通过提取、标注和序列化PubMed Central Open Access子集的全部内容,创建了一个包含超过2.4亿个唯一图像文本对的综合档案。框架还提供元数据和专业指导的注释。通过发布在BIOMEDICA数据集上持续预训练的BMCA-CLIP模型套件,展示了资源的实用性和可访问性。这些模型在多个任务上表现出卓越的性能,包括病理学、放射学、眼科、皮肤科、手术、分子生物学、寄生虫学和细胞生物学等领域,并且在零样本分类和图像文本检索方面表现出色。为促进可重复性和合作,向广大研究社区发布代码库和数据集。
Key Takeaways
- BIOMEDICA框架旨在解决生物医学视觉语言模型发展的数据限制问题。
- 该框架通过提取、标注和序列化PubMed Central Open Access子集的内容,创建了一个大规模且多样的数据集。
- BIOMEDICA数据集包含超过2.4亿个独特的图像文本对,涵盖广泛的生物医学知识。
- BMCA-CLIP模型套件在多个生物医学任务上表现出卓越性能,特别是在零样本分类方面有明显改进。
- BMCA-CLIP模型预训练过程中无需下载大量数据,计算效率显著提高。
- BIOMEDICA框架和数据集被公开发布,以促进可重复性和合作。
点此查看论文截图


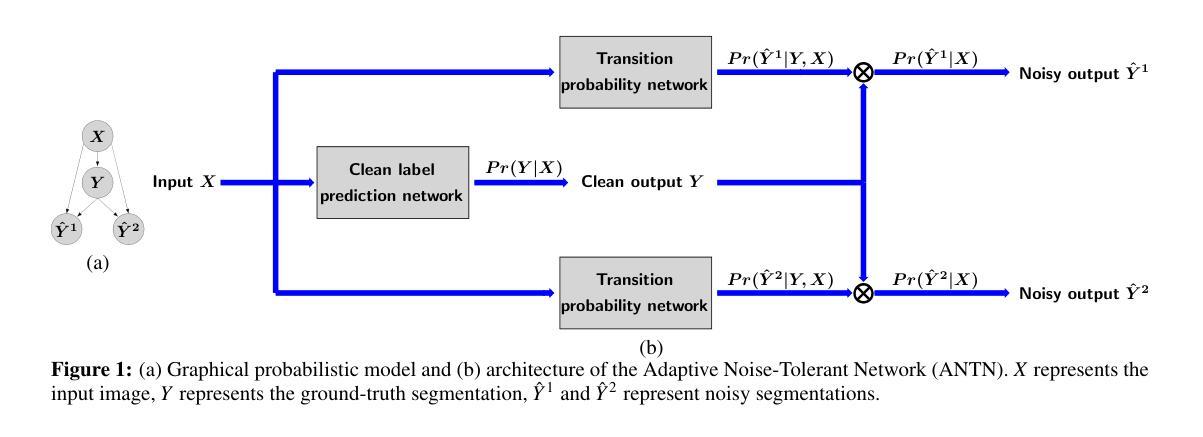
Adaptive Noise-Tolerant Network for Image Segmentation
Authors:Weizhi Li
Unlike image classification and annotation, for which deep network models have achieved dominating superior performances compared to traditional computer vision algorithms, deep learning for automatic image segmentation still faces critical challenges. One of such hurdles is to obtain ground-truth segmentations as the training labels for deep network training. Especially when we study biomedical images, such as histopathological images (histo-images), it is unrealistic to ask for manual segmentation labels as the ground truth for training due to the fine image resolution as well as the large image size and complexity. In this paper, instead of relying on clean segmentation labels, we study whether and how integrating imperfect or noisy segmentation results from off-the-shelf segmentation algorithms may help achieve better segmentation results through a new Adaptive Noise-Tolerant Network (ANTN) model. We extend the noisy label deep learning to image segmentation with two novel aspects: (1) multiple noisy labels can be integrated into one deep learning model; (2) noisy segmentation modeling, including probabilistic parameters, is adaptive, depending on the given testing image appearance. Implementation of the new ANTN model on both the synthetic data and real-world histo-images demonstrates its effectiveness and superiority over off-the-shelf and other existing deep-learning-based image segmentation algorithms.
与图像分类和标注不同,深度学习在自动图像分割方面仍然面临重大挑战,尽管深度网络模型在传统计算机视觉算法上取得了主导优势。其中一大挑战是获取真实分割作为深度网络训练的训练标签。特别是在研究生物医学图像(如组织病理学图像)时,由于图像分辨率高、尺寸大且复杂,要求手动分割标签作为训练的真实依据是不现实的。
本文不依赖于干净的分割标签,而是研究将现成的分割算法产生的有缺陷或嘈杂的分割结果集成起来,是否以及如何能够通过一种新的自适应噪声容忍网络(ANTN)模型,实现更好的分割结果。我们将噪声标签深度学习扩展到图像分割,并带来了两个新的观点:(1)多个噪声标签可以集成到一个深度学习模型中;(2)噪声分割建模,包括概率参数,是自适应的,取决于给定的测试图像外观。ANTN模型在合成数据和真实世界组织病理学图像上的实现,证明了其有效性,并优于现成的以及其他现有的基于深度学习的图像分割算法。
论文及项目相关链接
Summary
自动图像分割是深度学习中的一个挑战,因为获取用于深度网络训练的精确分割标签非常困难。针对生物医学图像等场景,本文提出了一种自适应噪声容忍网络(ANTN)模型,通过集成现成的分割算法得到的不完美或噪声分割结果,提高分割性能。该模型将噪声标签深度学习扩展到图像分割领域,具有两个创新点:一是能将多个噪声标签集成到一个深度学习模型中;二是根据给定的测试图像外观进行自适应的概率参数建模。实验表明,该模型在合成数据和真实世界历史图像上的表现优于现成的和其他基于深度学习的图像分割算法。
Key Takeaways
- 深度学习在自动图像分割上仍面临挑战,特别是获取用于深度网络训练的精确分割标签的难度大。
- 手动为生物医学图像(如组织病理学图像)制作精确的分割标签不切实际。
- 本文提出了自适应噪声容忍网络(ANTN)模型,能够集成不完美或噪声分割结果,提高图像分割性能。
- ANTN模型将多个噪声标签集成到深度学习模型中,并实现自适应的概率参数建模。
点此查看论文截图

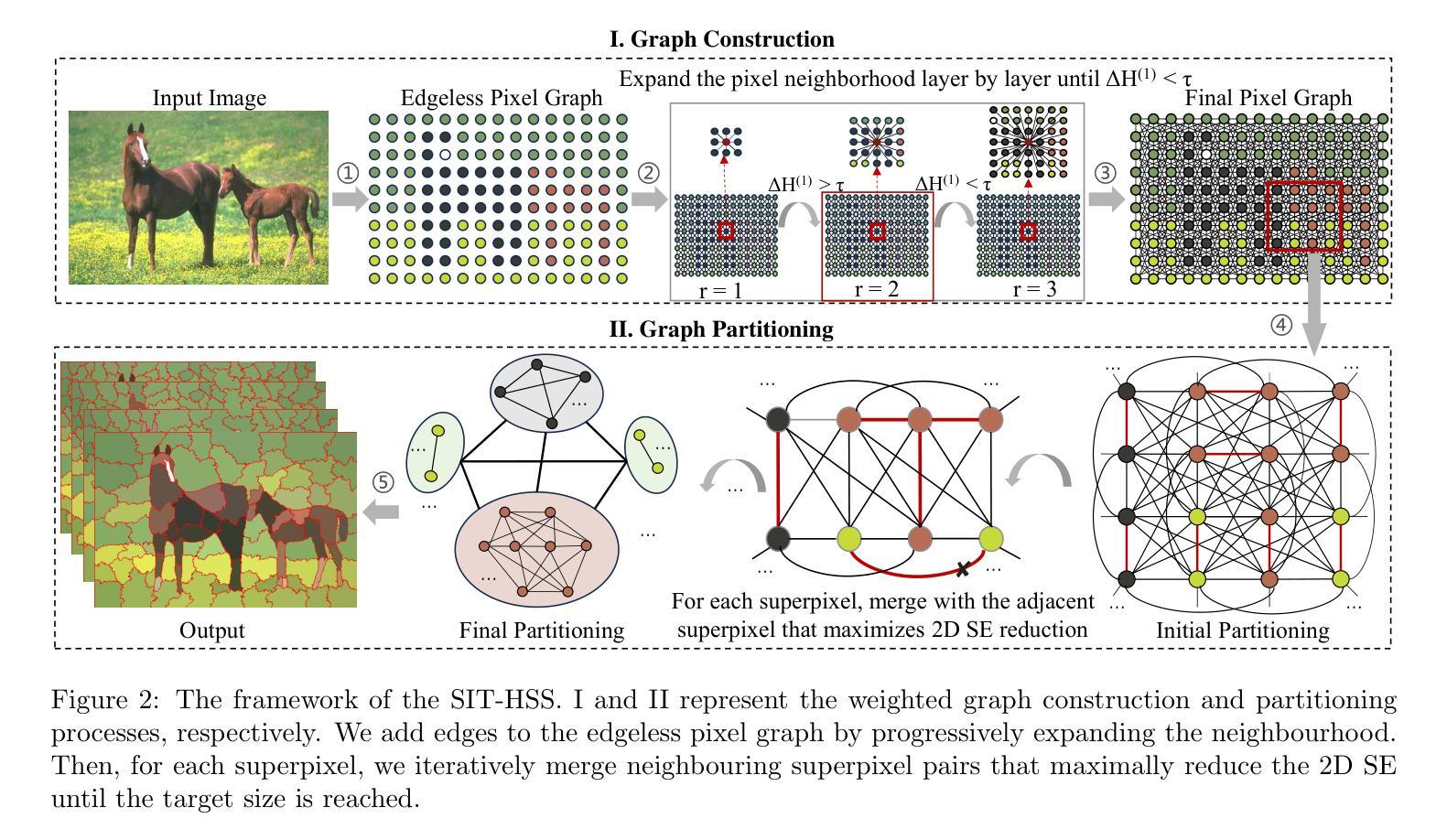
Hierarchical Superpixel Segmentation via Structural Information Theory
Authors:Minhui Xie, Hao Peng, Pu Li, Guangjie Zeng, Shuhai Wang, Jia Wu, Peng Li, Philip S. Yu
Superpixel segmentation is a foundation for many higher-level computer vision tasks, such as image segmentation, object recognition, and scene understanding. Existing graph-based superpixel segmentation methods typically concentrate on the relationships between a given pixel and its directly adjacent pixels while overlooking the influence of non-adjacent pixels. These approaches do not fully leverage the global information in the graph, leading to suboptimal segmentation quality. To address this limitation, we present SIT-HSS, a hierarchical superpixel segmentation method based on structural information theory. Specifically, we first design a novel graph construction strategy that incrementally explores the pixel neighborhood to add edges based on 1-dimensional structural entropy (1D SE). This strategy maximizes the retention of graph information while avoiding an overly complex graph structure. Then, we design a new 2D SE-guided hierarchical graph partitioning method, which iteratively merges pixel clusters layer by layer to reduce the graph’s 2D SE until a predefined segmentation scale is achieved. Experimental results on three benchmark datasets demonstrate that the SIT-HSS performs better than state-of-the-art unsupervised superpixel segmentation algorithms. The source code is available at \url{https://github.com/SELGroup/SIT-HSS}.
超像素分割是许多高级计算机视觉任务的基础,如图像分割、对象识别和场景理解。现有的基于图的超像素分割方法通常侧重于给定像素及其直接相邻像素之间的关系,而忽略了非相邻像素的影响。这些方法没有充分利用图中的全局信息,导致分割质量不佳。为了解决这一局限性,我们提出了基于结构信息理论的分层超像素分割方法(简称SIT-HSS)。具体来说,我们首先设计了一种新的图构建策略,该策略基于一维结构熵(1D SE)增量地探索像素邻域以添加边。这种策略最大限度地保留了图形信息,同时避免了过于复杂的图形结构。然后,我们设计了一种新的由二维SE引导的分层次图划分方法,该方法逐层迭代合并像素簇,以减少图的二维SE,直到达到预定的分割尺度。在三个基准数据集上的实验结果表明,与最新的无监督超像素分割算法相比,SIT-HSS具有更好的性能。源代码可在https://github.com/SELGroup/SIT-HSS获取。
论文及项目相关链接
PDF Accepted by SDM 2025
Summary
基于结构信息理论的分层超像素分割方法,通过设计新的图构建策略,基于一维结构熵(1D SE)增量探索像素邻域来添加边缘,并设计新的二维SE引导分层图划分方法,实现图像超像素分割,优于现有先进无监督超像素分割算法。
Key Takeaways
- 超像素分割是计算机视觉任务的基础,如图像分割、目标识别和场景理解。
- 现有基于图的超像素分割方法主要关注给定像素及其直接相邻像素之间的关系,忽略了非相邻像素的影响。
- 现有方法没有充分利用图中的全局信息,导致分割质量不佳。
- SIT-HSS是一种基于结构信息理论的分层超像素分割方法,旨在解决上述问题。
- SIT-HSS设计了一种新的图构建策略,该策略基于一维结构熵(1D SE)增量探索像素邻域来添加边缘。
- SIT-HSS还设计了一种新的二维SE引导分层图划分方法,通过逐层合并像素簇来减少图的二维SE,直至达到预设的分割尺度。
点此查看论文截图

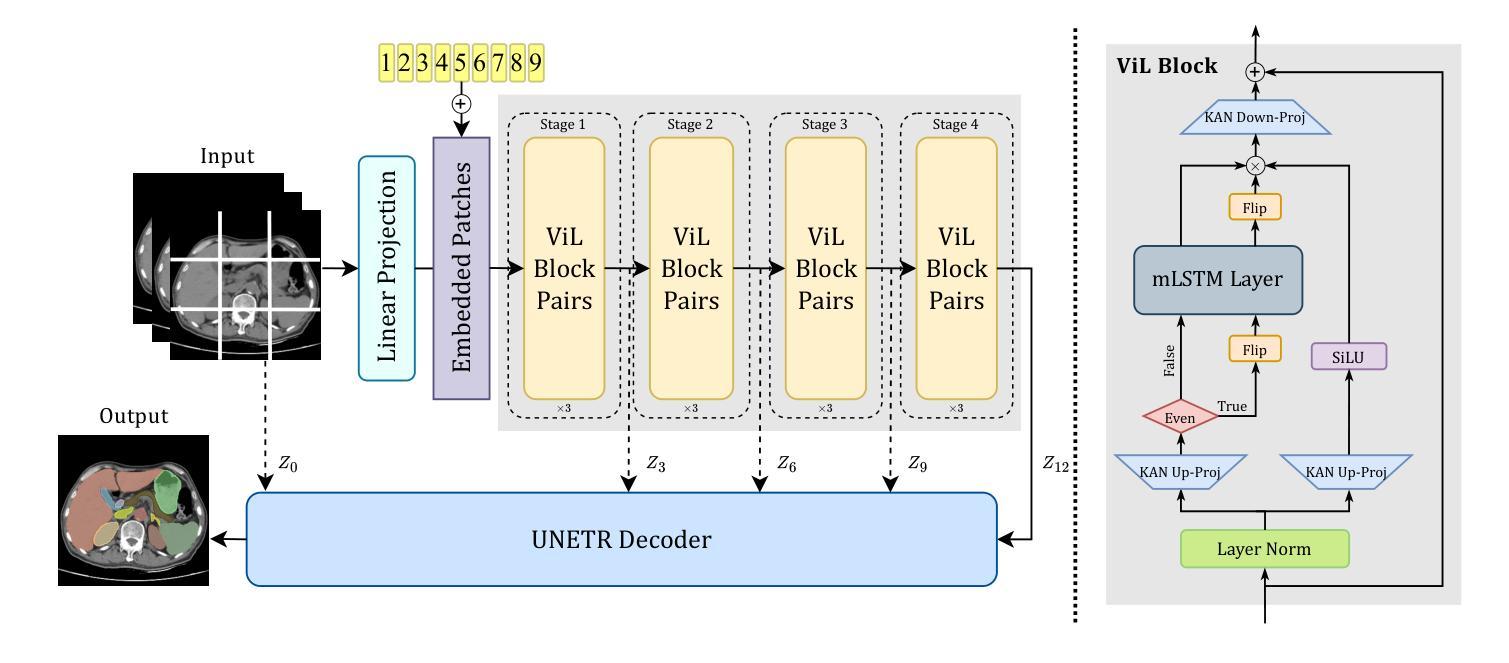
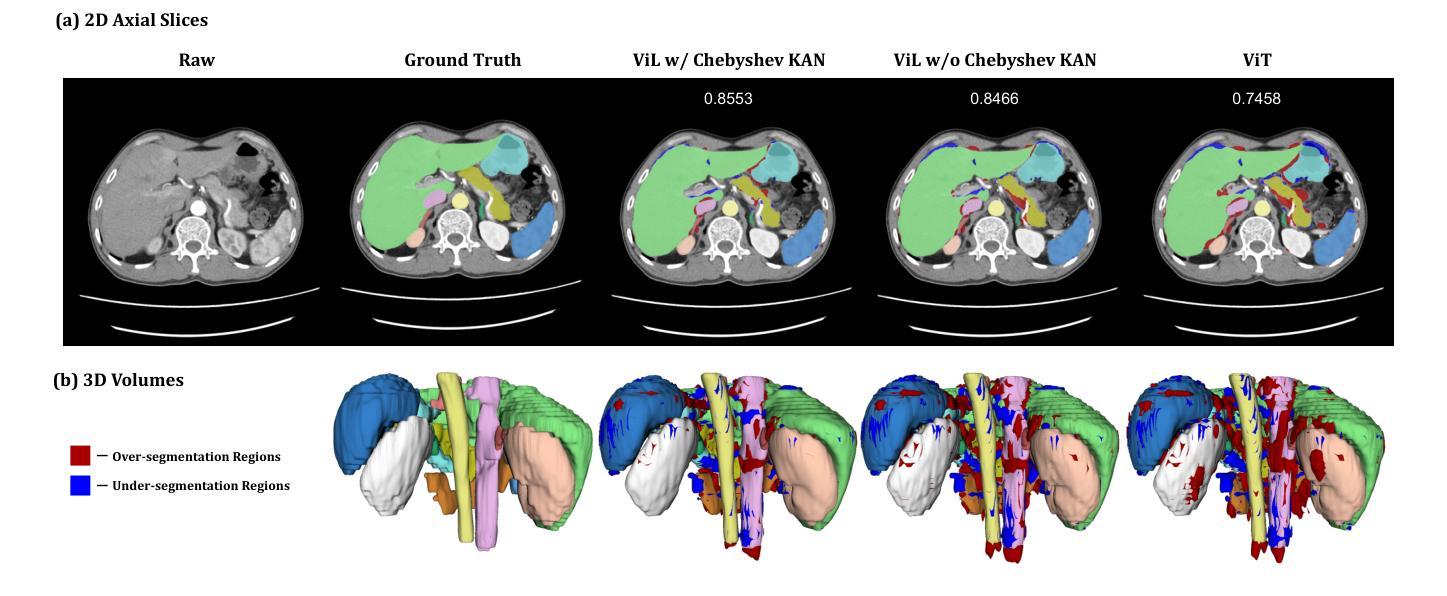
UNetVL: Enhancing 3D Medical Image Segmentation with Chebyshev KAN Powered Vision-LSTM
Authors:Xuhui Guo, Tanmoy Dam, Rohan Dhamdhere, Gourav Modanwal, Anant Madabhushi
3D medical image segmentation has progressed considerably due to Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs), yet these methods struggle to balance long-range dependency acquisition with computational efficiency. To address this challenge, we propose UNETVL (U-Net Vision-LSTM), a novel architecture that leverages recent advancements in temporal information processing. UNETVL incorporates Vision-LSTM (ViL) for improved scalability and memory functions, alongside an efficient Chebyshev Kolmogorov-Arnold Networks (KAN) to handle complex and long-range dependency patterns more effectively. We validated our method on the ACDC and AMOS2022 (post challenge Task 2) benchmark datasets, showing a significant improvement in mean Dice score compared to recent state-of-the-art approaches, especially over its predecessor, UNETR, with increases of 7.3% on ACDC and 15.6% on AMOS, respectively. Extensive ablation studies were conducted to demonstrate the impact of each component in UNETVL, providing a comprehensive understanding of its architecture. Our code is available at https://github.com/tgrex6/UNETVL, facilitating further research and applications in this domain.
三维医学图像分割领域已经由于卷积神经网络(CNN)和视觉转换器(ViT)的发展取得了显著的进步。然而,这些方法在平衡长距离依赖获取与计算效率方面仍存在挑战。为了应对这一挑战,我们提出了UNETVL(U-Net Vision-LSTM)这一新型架构,该架构利用最新的时间信息处理进展。UNETVL结合了Vision-LSTM(ViL)以增强其可扩展性和记忆功能,并采用了高效的切比雪夫Kolmogorov-Arnold网络(KAN)以更有效地处理复杂且长距离的依赖模式。我们在ACDC和AMOS2022(挑战赛任务2之后)的基准数据集上验证了我们的方法,相较于最新的最先进方法,特别是在其前身UNETR上,我们的方法在ACDC和AMOS上的平均Dice得分均显著提高,分别提高了7.3%和15.6%。进行了广泛的消融研究,以展示UNETVL中每个组件的影响,为其架构提供了全面的理解。我们的代码可通过https://github.com/tgrex6/UNETVL获取,为这一领域的进一步研究和应用提供了便利。
论文及项目相关链接
Summary
这篇论文介绍了新的架构UNETVL,该架构结合了卷积神经网络(CNN)、视觉转换器(ViT)和时空信息处理的最新进展来解决医学图像分割问题。它通过引入Vision-LSTM(ViL)提高了可扩展性和内存功能,并使用高效的切比雪夫柯尔莫哥洛夫-阿诺尔德网络(KAN)更有效地处理复杂和长期的依赖模式。实验结果显示,该架构在ACDC和AMOS2022基准数据集上的表现显著优于最新的先进方法,特别是与前身的UNET相比,Dice系数的平均得分分别提高了7.3%和增加了两倍。文章提供了完整的UNETVL代码供公众参考,以便在此领域进行进一步的研究和应用。这一设计可为复杂医疗图像处理任务提供更高效和准确的技术手段。
Key Takeaways
以下是基于上述文本的重要见解列表:
- UNETVL是一个结合了卷积神经网络(CNN)、视觉转换器(ViT)和时空信息处理的最新进展的医学图像分割架构。它旨在解决长期依赖获取与计算效率之间的平衡问题。
- UNETVL引入了Vision-LSTM(ViL),旨在提高模型的内存功能和可扩展性。它通过更高效处理长期依赖模式改进了模型的性能。
- 切比雪夫柯尔莫哥洛夫-阿诺尔德网络(KAN)被用于UNETVL中,以更有效地处理复杂的长期依赖关系模式。这为模型提供了更好的性能提升。
点此查看论文截图


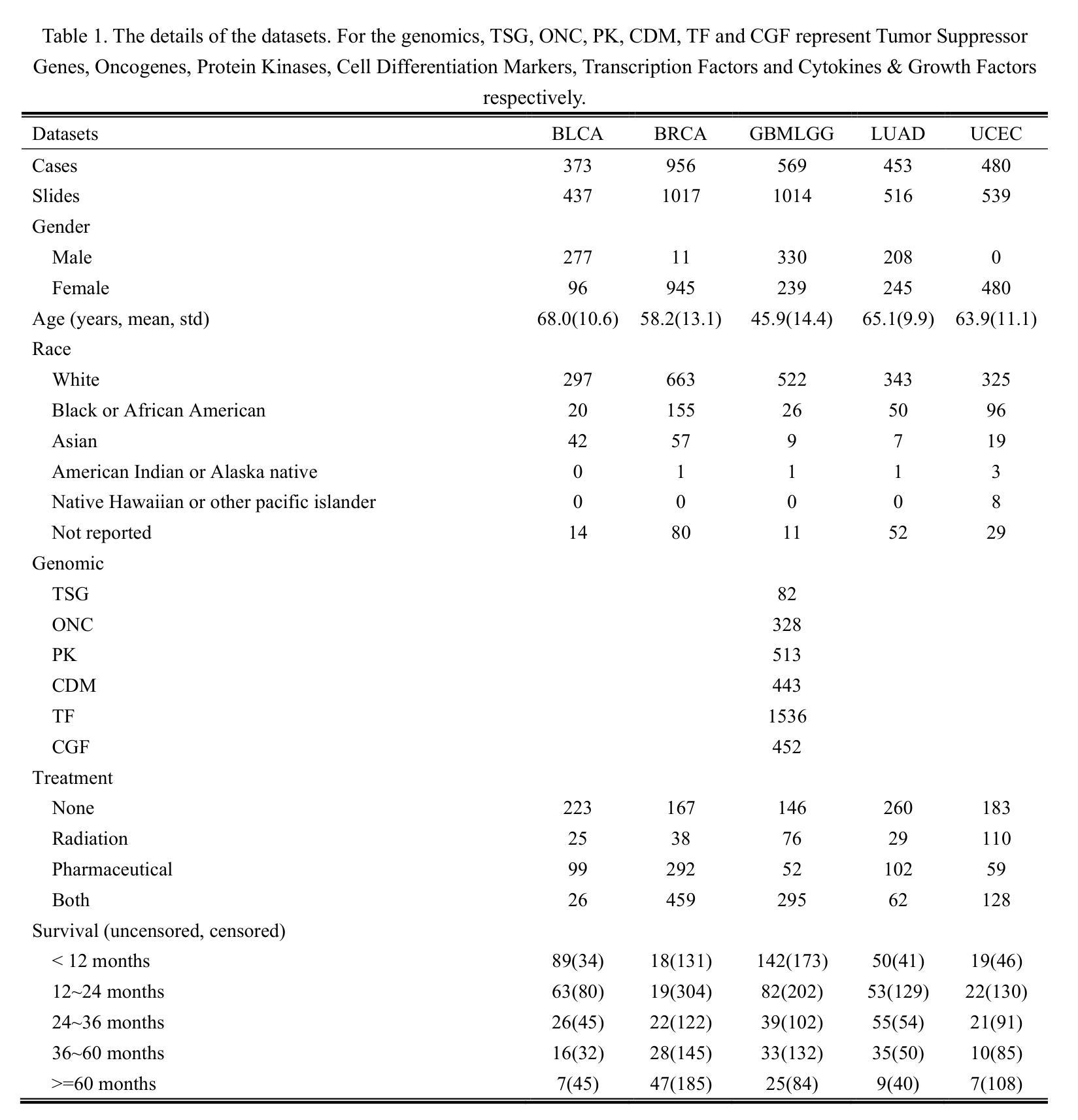
A Multi-Modal Deep Learning Framework for Pan-Cancer Prognosis
Authors:Binyu Zhang, Shichao Li, Junpeng Jian, Zhu Meng, Limei Guo, Zhicheng Zhao
Prognostic task is of great importance as it closely related to the survival analysis of patients, the optimization of treatment plans and the allocation of resources. The existing prognostic models have shown promising results on specific datasets, but there are limitations in two aspects. On the one hand, they merely explore certain types of modal data, such as patient histopathology WSI and gene expression analysis. On the other hand, they adopt the per-cancer-per-model paradigm, which means the trained models can only predict the prognostic effect of a single type of cancer, resulting in weak generalization ability. In this paper, a deep-learning based model, named UMPSNet, is proposed. Specifically, to comprehensively understand the condition of patients, in addition to constructing encoders for histopathology images and genomic expression profiles respectively, UMPSNet further integrates four types of important meta data (demographic information, cancer type information, treatment protocols, and diagnosis results) into text templates, and then introduces a text encoder to extract textual features. In addition, the optimal transport OT-based attention mechanism is utilized to align and fuse features of different modalities. Furthermore, a guided soft mixture of experts (GMoE) mechanism is introduced to effectively address the issue of distribution differences among multiple cancer datasets. By incorporating the multi-modality of patient data and joint training, UMPSNet outperforms all SOTA approaches, and moreover, it demonstrates the effectiveness and generalization ability of the proposed learning paradigm of a single model for multiple cancer types. The code of UMPSNet is available at https://github.com/binging512/UMPSNet.
预后任务非常重要,因为它与患者的生存分析、治疗计划的优化和资源的分配密切相关。现有的预后模型在特定数据集上取得了有前景的结果,但有两个方面的局限性。一方面,它们仅仅探索了某些类型的模态数据,如患者的病理组织学WSI和基因表达分析。另一方面,它们采用了一种针对每种癌症的模型模式,这意味着训练过的模型只能预测单一类型癌症的预后效果,导致泛化能力较弱。本文提出了一种基于深度学习的模型,名为UMPSNet。具体来说,为了全面理解患者的病情,除了分别为病理组织学图像和基因表达谱构建编码器外,UMPSNet还进一步将四种重要的元数据(人口统计信息、癌症类型信息、治疗协议和诊断结果)整合到文本模板中,然后引入一个文本编码器来提取文本特征。此外,还利用基于最优传输的注意力机制来对齐和融合不同模态的特征。此外,还引入了导向软混合专家(GMoE)机制,有效解决多个癌症数据集之间分布差异的问题。通过结合患者数据的多模态联合训练,UMPSNet优于所有最先进的方法,而且验证了单一模型用于多种癌症类型的学习模式的的有效性和泛化能力。UMPSNet的代码可在https://github.com/binging512/UMPSNet获取。
论文及项目相关链接
Summary
本文提出一种基于深度学习的多模态融合模型UMPSNet,旨在更全面地了解病人情况并预测多类型癌症的预后效果。模型融合了病人的四种重要元数据,采用文本编码器提取文本特征,并利用最优传输的注意力机制对齐和融合不同模态的特征。此外,引入引导软混合专家机制解决多个癌症数据集间的分布差异问题。该模型展现出优越的性能和泛化能力。
Key Takeaways
- UMPSNet模型是一个基于深度学习的多模态融合模型,用于病人的全面情况理解和多类型癌症的预后效果预测。
- 该模型融合了病人的四种重要元数据,包括人口学信息、癌症类型信息、治疗协议和诊断结果,并转化为文本模板进行处理。
- UMPSNet采用文本编码器提取文本特征,并应用最优传输的注意力机制来对齐和融合不同模态的特征。
- 引导软混合专家机制被引入以解决多个癌症数据集间的分布差异问题。
- UMPSNet模型展现出优越的性能,优于当前所有先进方法,并验证了单一模型对多种癌症类型学习的有效性和泛化能力。
- UMPSNet模型的代码已公开可获取。
点此查看论文截图

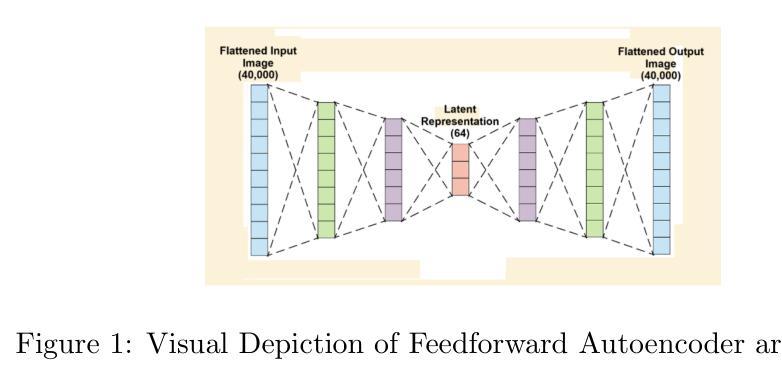
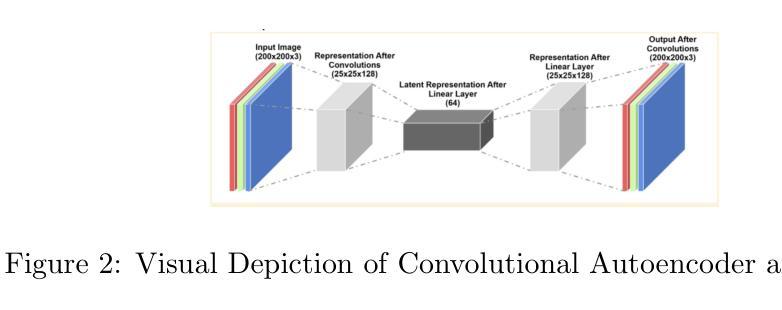
Comparison of Autoencoders for tokenization of ASL datasets
Authors:Vouk Praun-Petrovic, Aadhvika Koundinya, Lavanya Prahallad
Generative AI, powered by large language models (LLMs), has revolutionized applications across text, audio, images, and video. This study focuses on developing and evaluating encoder-decoder architectures for the American Sign Language (ASL) image dataset, consisting of 87,000 images across 29 hand sign classes. Three approaches were compared: Feedforward Autoencoders, Convolutional Autoencoders, and Diffusion Autoencoders. The Diffusion Autoencoder outperformed the others, achieving the lowest mean squared error (MSE) and highest Mean Opinion Score (MOS) due to its probabilistic noise modeling and iterative denoising capabilities. The Convolutional Autoencoder demonstrated effective spatial feature extraction but lacked the robustness of the diffusion process, while the Feedforward Autoencoder served as a baseline with limitations in handling complex image data. Objective and subjective evaluations confirmed the superiority of the Diffusion Autoencoder for high-fidelity image reconstruction, emphasizing its potential in multimodal AI applications such as sign language recognition and generation. This work provides critical insights into designing robust encoder-decoder systems to advance multimodal AI capabilities.
生成式人工智能(Generative AI)凭借大型语言模型(LLMs)的力量,在文本、音频、图像和视频等领域的应用中引发了革命性的变革。本研究专注于为美国手语(ASL)图像数据集开发并评估编码器-解码器架构。该数据集包含跨29个手语类别的87,000张图像。研究比较了三种方法:前馈自编码器(Feedforward Autoencoders)、卷积自编码器(Convolutional Autoencoders)和扩散自编码器(Diffusion Autoencoders)。由于具有概率噪声建模和迭代去噪能力,扩散自编码器在性能上优于其他方法,取得了最低的平均平方误差(MSE)和最高的平均意见分数(MOS)。卷积自编码器显示出有效的空间特征提取能力,但在扩散过程的稳健性方面存在不足,而前馈自编码器则作为处理复杂图像数据存在局限性的基线模型。客观和主观评估证实了扩散自编码器在高保真图像重建方面的优越性,强调其在手语识别和生成等多模态人工智能应用中的潜力。本研究为设计稳健的编码器-解码器系统提供了关键见解,有助于推动多模态人工智能能力的发展。
论文及项目相关链接
PDF 9 pages, 2 tables, 4 figures
Summary
基于大型语言模型(LLM)的生成式AI已经彻底改变了文本、音频、图像和视频的应用方式。本研究专注于针对美国手语(ASL)图像数据集开发和评估编码器-解码器架构,包含87,000张跨越29个手势类别的图像。对比了三种方法:前馈自编码器、卷积自编码器和扩散自编码器。扩散自编码器因概率噪声建模和迭代去噪能力表现最佳,实现了最低均方误差(MSE)和最高平均意见得分(MOS)。卷积自编码器在提取空间特征方面表现出色,但缺乏扩散过程的稳健性;而前馈自编码器作为基准,在处理复杂图像数据时存在局限性。客观和主观评估证实了扩散自编码器在高保真图像重建中的优越性,强调其在手势语言识别和生成等多模式AI应用中的潜力。本研究为设计稳健的编码器-解码器系统以推动多模式AI能力提供了关键见解。
Key Takeaways
- 生成式AI借助大型语言模型(LLM)已广泛应用于文本、音频、图像和视频领域。
- 本研究聚焦于针对美国手语(ASL)图像数据集开发和评估编码器-解码器架构。
- 对比了三种图像处理方法:前馈自编码器、卷积自编码器和扩散自编码器。
- 扩散自编码器因概率噪声建模和迭代去噪能力表现最佳。
- 卷积自编码器在提取空间特征方面表现出色,但缺乏扩散稳健性。
- 高保真图像重建中扩散自编码器的优越性通过客观和主观评估得到证实。
点此查看论文截图

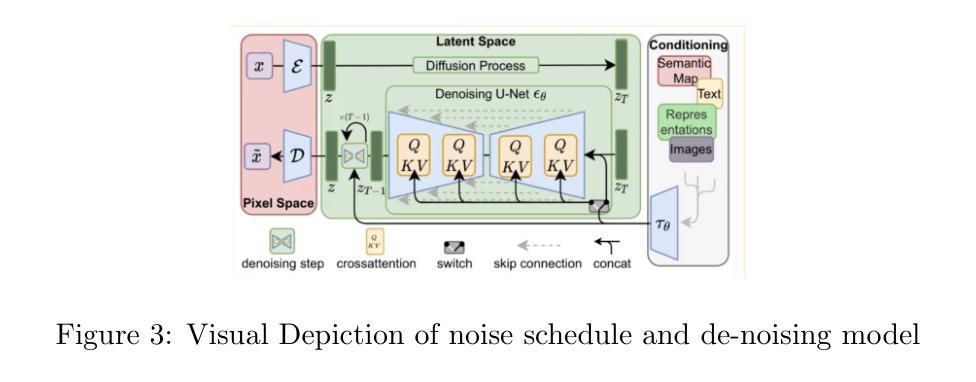

A Foundational Generative Model for Breast Ultrasound Image Analysis
Authors:Haojun Yu, Youcheng Li, Nan Zhang, Zihan Niu, Xuantong Gong, Yanwen Luo, Haotian Ye, Siyu He, Quanlin Wu, Wangyan Qin, Mengyuan Zhou, Jie Han, Jia Tao, Ziwei Zhao, Di Dai, Di He, Dong Wang, Binghui Tang, Ling Huo, James Zou, Qingli Zhu, Yong Wang, Liwei Wang
Foundational models have emerged as powerful tools for addressing various tasks in clinical settings. However, their potential development to breast ultrasound analysis remains untapped. In this paper, we present BUSGen, the first foundational generative model specifically designed for breast ultrasound image analysis. Pretrained on over 3.5 million breast ultrasound images, BUSGen has acquired extensive knowledge of breast structures, pathological features, and clinical variations. With few-shot adaptation, BUSGen can generate repositories of realistic and informative task-specific data, facilitating the development of models for a wide range of downstream tasks. Extensive experiments highlight BUSGen’s exceptional adaptability, significantly exceeding real-data-trained foundational models in breast cancer screening, diagnosis, and prognosis. In breast cancer early diagnosis, our approach outperformed all board-certified radiologists (n=9), achieving an average sensitivity improvement of 16.5% (P-value<0.0001). Additionally, we characterized the scaling effect of using generated data which was as effective as the collected real-world data for training diagnostic models. Moreover, extensive experiments demonstrated that our approach improved the generalization ability of downstream models. Importantly, BUSGen protected patient privacy by enabling fully de-identified data sharing, making progress forward in secure medical data utilization. An online demo of BUSGen is available at https://aibus.bio.
基础模型已作为临床环境中处理各种任务的强大工具而出现。然而,它们在乳腺超声分析方面的潜力尚未被开发。在本文中,我们提出了BUSGen,这是专门为乳腺超声图像分析设计的首个基础生成模型。经过在超过350万张乳腺超声图像上的预训练,BUSGen获得了关于乳房结构、病理特征和临床变化的广泛知识。通过少量的适应,BUSGen可以生成实际且信息量丰富的特定任务数据仓库,促进针对各种下游任务的模型发展。大量实验突出了BUSGen的出色适应性,在乳腺癌筛查、诊断和预后方面显著超过了使用真实数据训练的基础模型。在乳腺癌早期诊断中,我们的方法优于所有执业医师认证的放射科医生(n=9),平均敏感性提高了16.5%(P值<0.0001)。此外,我们还分析了使用生成数据与收集的真实世界数据训练诊断模型的规模效应。而且,大量实验证明,我们的方法提高了下游模型的泛化能力。重要的是,BUSGen通过实现完全匿名化的数据共享,保护了患者隐私,在安全利用医疗数据方面取得了进展。BUSGen的在线演示可在https://aibus.bio上找到。
论文及项目相关链接
PDF Peking University; Stanford University; Peking University Cancer Hospital & Institute; Peking Union Medical College Hospital; Cancer Hospital, Chinese Academy of Medical Sciences
Summary
BUSGen是第一款专为乳腺超声图像分析设计的通用生成模型。该模型预训练在超过3.5万例乳腺超声图像上,并具备强大的泛化能力。它能够在少量样本适应下生成现实且具有信息性的任务特定数据,促进了各种下游任务模型的发展。对于乳腺癌筛查、诊断及预后等方面,BUSGen的表现极为出色,其适应性显著超过了由真实数据训练出的通用模型。此外,该模型还能有效保护患者隐私,推动安全医疗数据的利用。目前可通过https://aibus.bio在线体验。
Key Takeaways
- BUSGen是专为乳腺超声图像分析设计的首款通用生成模型。
- 预训练在大量乳腺超声图像上,涵盖广泛的乳腺结构、病理特征和临床变化知识。
- 通过少量样本适应,能生成具有现实性和信息性的任务特定数据。
- 在乳腺癌筛查、诊断和预后方面表现优异,超越了许多真实数据训练的通用模型。
- 有效提高下游模型的泛化能力。
- 保护患者隐私,推动安全医疗数据的利用。
点此查看论文截图

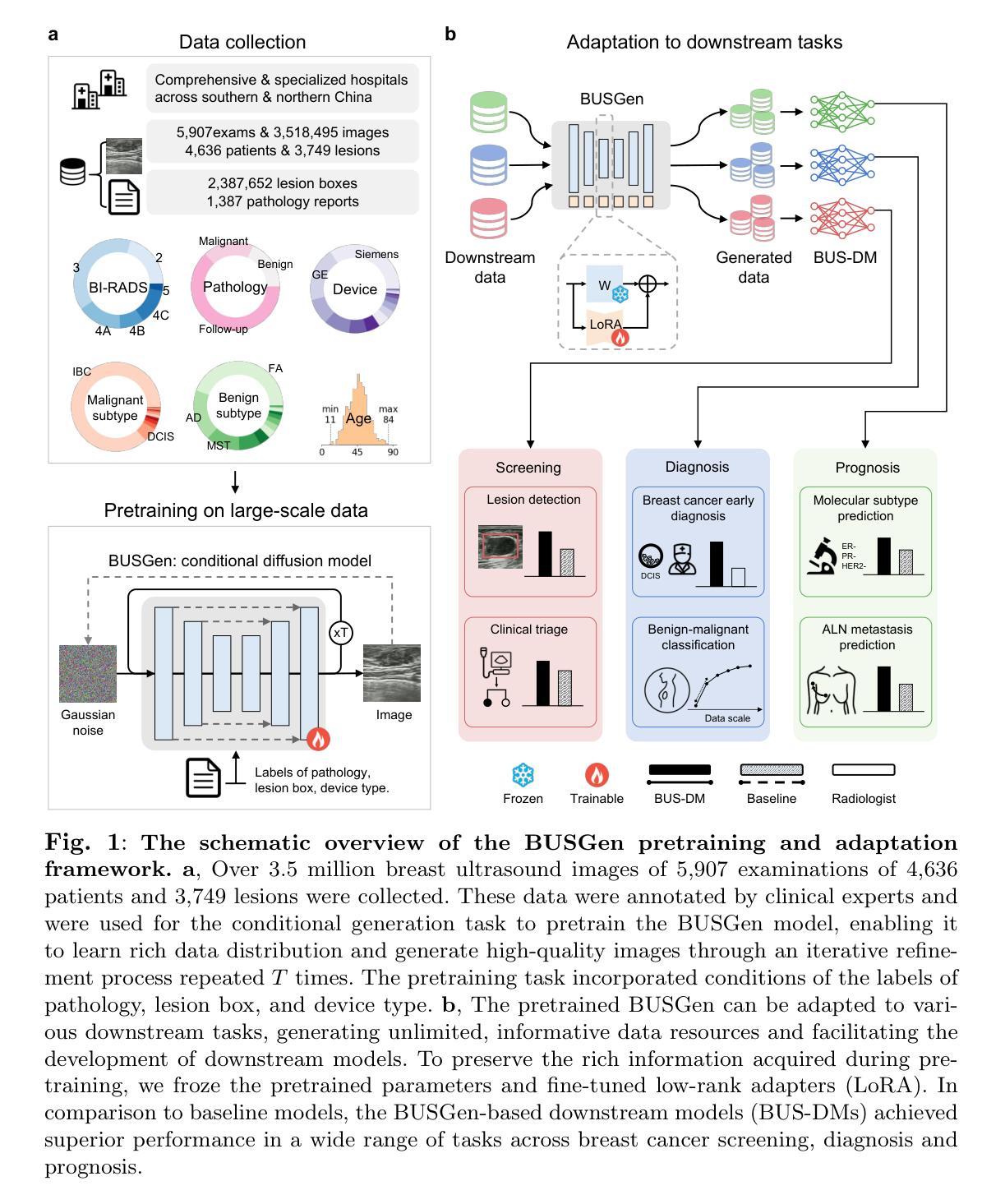
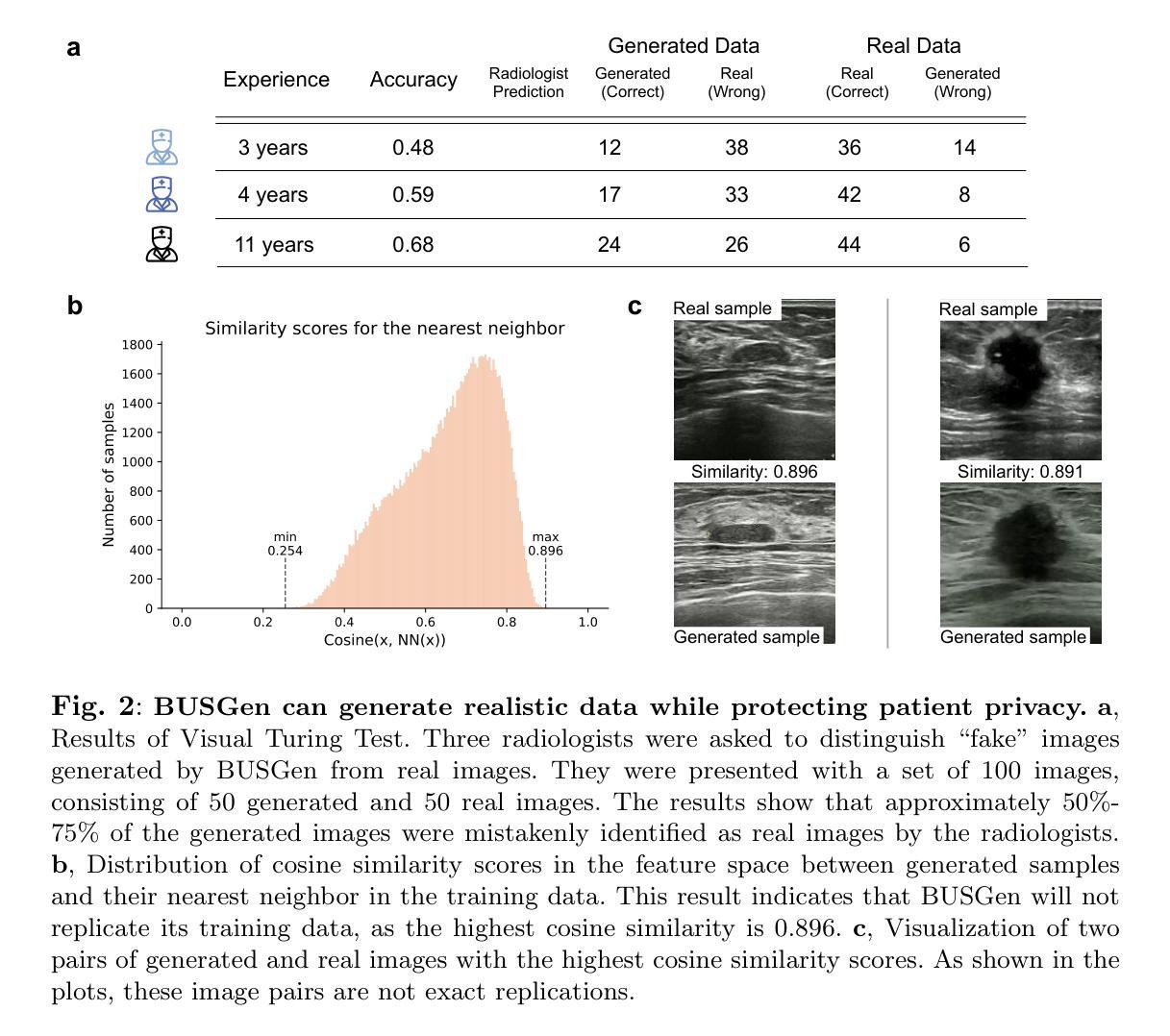
UR2P-Dehaze: Learning a Simple Image Dehaze Enhancer via Unpaired Rich Physical Prior
Authors:Minglong Xue, Shuaibin Fan, Shivakumara Palaiahnakote, Mingliang Zhou
Image dehazing techniques aim to enhance contrast and restore details, which are essential for preserving visual information and improving image processing accuracy. Existing methods rely on a single manual prior, which cannot effectively reveal image details. To overcome this limitation, we propose an unpaired image dehazing network, called the Simple Image Dehaze Enhancer via Unpaired Rich Physical Prior (UR2P-Dehaze). First, to accurately estimate the illumination, reflectance, and color information of the hazy image, we design a shared prior estimator (SPE) that is iteratively trained to ensure the consistency of illumination and reflectance, generating clear, high-quality images. Additionally, a self-monitoring mechanism is introduced to eliminate undesirable features, providing reliable priors for image reconstruction. Next, we propose Dynamic Wavelet Separable Convolution (DWSC), which effectively integrates key features across both low and high frequencies, significantly enhancing the preservation of image details and ensuring global consistency. Finally, to effectively restore the color information of the image, we propose an Adaptive Color Corrector that addresses the problem of unclear colors. The PSNR, SSIM, LPIPS, FID and CIEDE2000 metrics on the benchmark dataset show that our method achieves state-of-the-art performance. It also contributes to the performance improvement of downstream tasks. The project code will be available at https://github.com/Fan-pixel/UR2P-Dehaze. \end{abstract}
图像去雾技术旨在提高对比度和恢复细节,这对于保留视觉信息、提高图像处理精度至关重要。现有方法依赖于单一的人工先验,无法有效揭示图像细节。为了克服这一局限性,我们提出了一种非配对图像去雾网络,称为通过非配对丰富物理先验的简单图像去雾增强器(UR2P-Dehaze)。首先,为了准确估计雾图像的照明、反射和颜色信息,我们设计了一个共享先验估计器(SPE),通过迭代训练确保照明和反射的一致性,生成清晰的高质量图像。此外,还引入了一种自我监控机制,以消除不良特征,为图像重建提供可靠的先验。接下来,我们提出了动态小波可分离卷积(DWSC),能够有效整合低频和高频的关键特征,显著增强图像细节的保留,并确保全局一致性。最后,为了有效恢复图像的颜色信息,我们提出了自适应颜色校正器,解决了颜色不清晰的问题。在基准数据集上的PSNR、SSIM、LPIPS、FID和CIEDE2000指标表明,我们的方法达到了最先进的性能。此外,它还为下游任务的性能改进做出了贡献。项目代码将在https://github.com/Fan-pixel/UR2P-Dehaze上提供。
论文及项目相关链接
摘要
本文主要介绍了一种名为UR2P-Dehaze的去雾网络。该网络旨在提高图像的对比度和细节恢复能力,通过设计共享先验估计器(SPE)和动态小波可分离卷积(DWSC)等技术手段,提高了去雾效果。此外,还引入了一种自适应颜色校正器来解决颜色不清晰的问题。在基准数据集上的PSNR、SSIM、LPIPS、FID和CIEDE2000等指标表明,该方法达到了最先进的性能,并能提高下游任务的性能。
关键见解
- 去雾技术旨在提高图像的对比度和细节恢复能力,对于保护视觉信息和提高图像处理精度至关重要。
- 现有方法依赖于单一的手动先验,无法有效揭示图像细节。
- UR2P-Dehaze网络通过设计共享先验估计器(SPE)来准确估计雾霾图像的照明、反射和颜色信息。
- 引入了一种自我监控机制,以消除不良特征,为图像重建提供可靠的先验。
- 动态小波可分离卷积(DWSC)能有效整合高低频的关键特征,增强图像细节的保护和全局一致性。
- UR2P-Dehaze通过自适应颜色校正器解决了颜色不清晰的问题。
- 在基准数据集上的多项指标表明,UR2P-Dehaze方法达到了最先进的性能,并能提高下游任务的性能。
点此查看论文截图

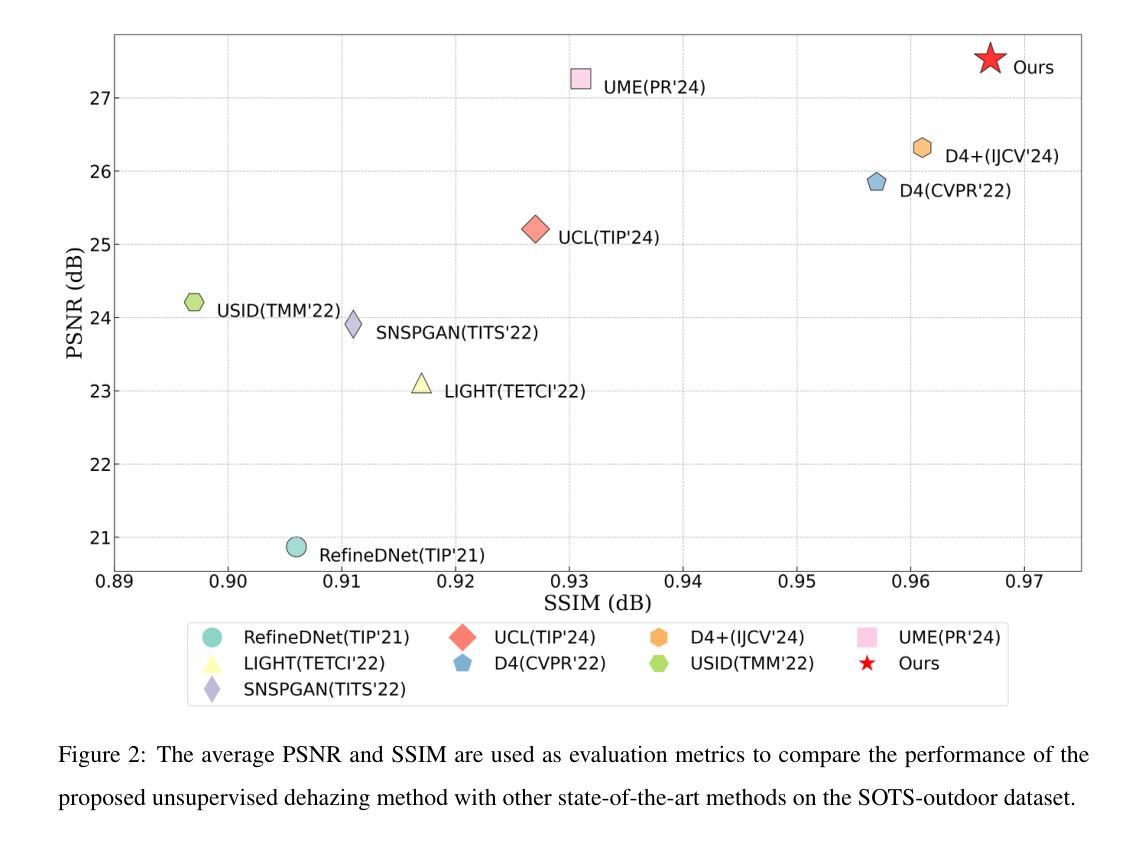
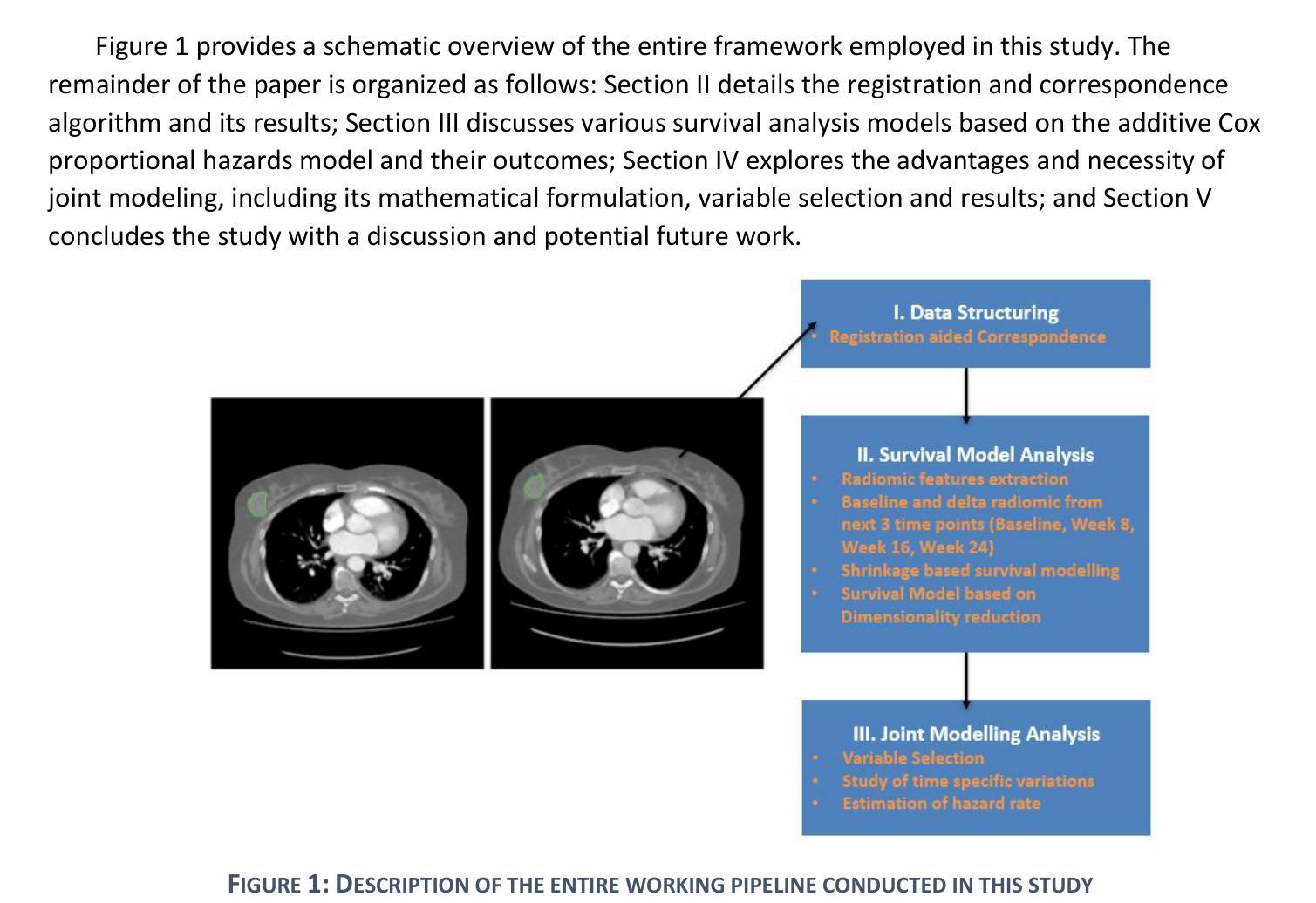
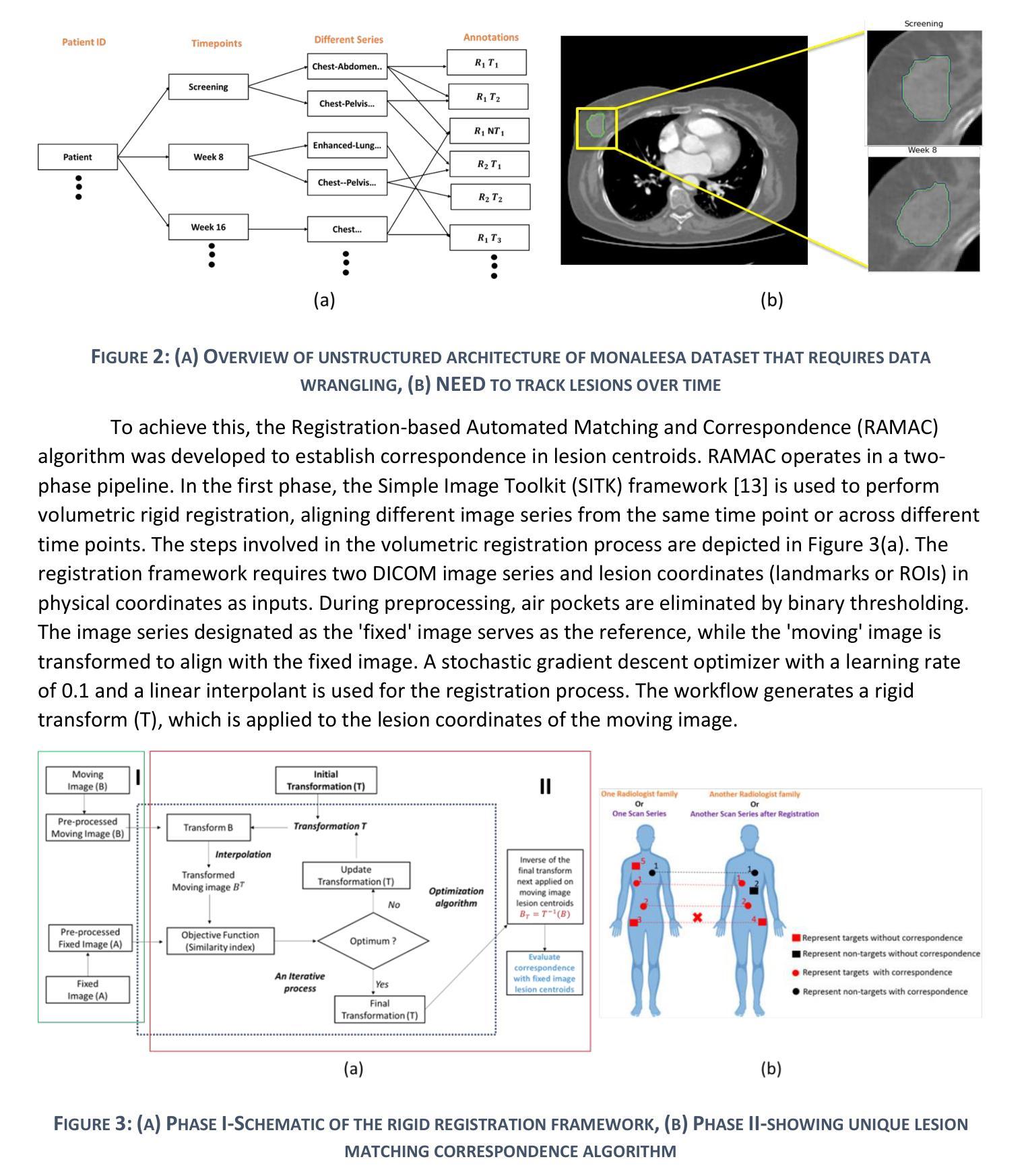
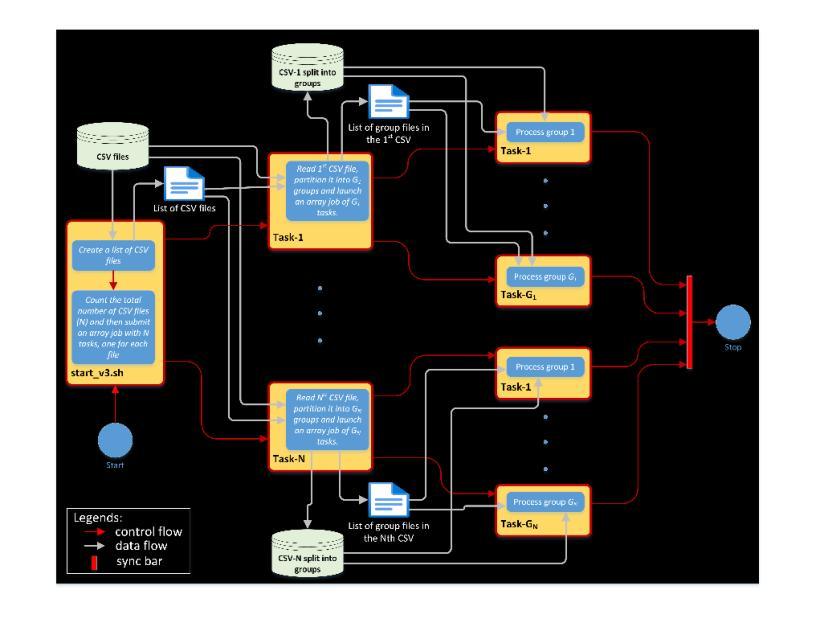
Improved joint modelling of breast cancer radiomics features and hazard by image registration aided longitudinal CT data
Authors:Subrata Mukherjee
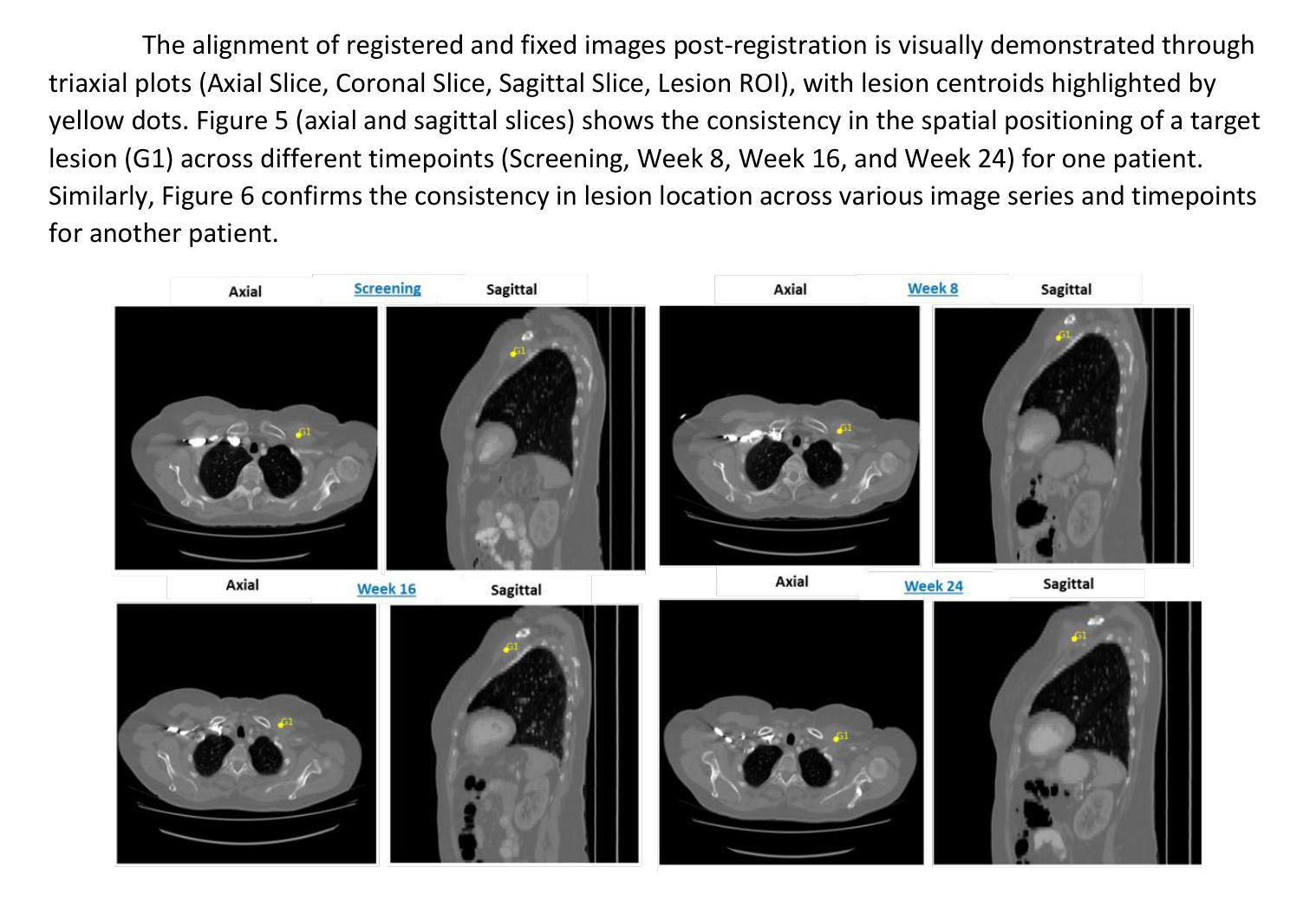
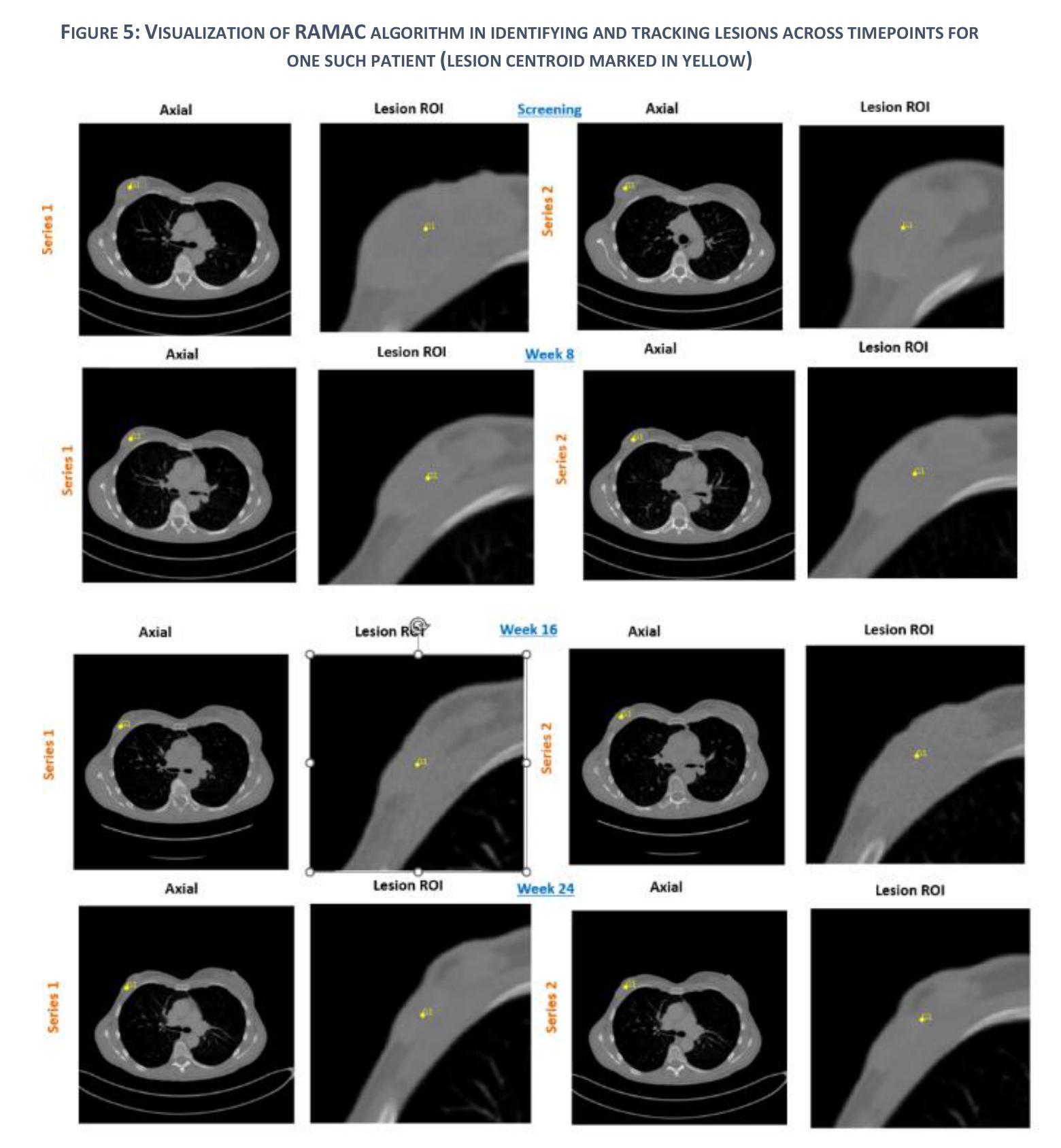
Patients with metastatic breast cancer (mBC) undergo continuous medical imaging during treatment, making accurate lesion detection and monitoring over time critical for clinical decisions. Predicting drug response from post-treatment data is essential for personalized care and pharmacological research. In collaboration with the U.S. Food and Drug Administration and Novartis Pharmaceuticals, we analyzed serial chest CT scans from two large-scale Phase III trials, MONALEESA 3 and MONALEESA 7. This paper has two objectives (a) Data Structuring developing a Registration Aided Automated Correspondence (RAMAC) algorithm for precise lesion tracking in longitudinal CT data, and (b) Survival Analysis creating imaging features and models from RAMAC structured data to predict patient outcomes. The RAMAC algorithm uses a two phase pipeline: three dimensional rigid registration aligns CT images, and a distance metric-based Hungarian algorithm tracks lesion correspondence. Using structured data, we developed interpretable models to assess progression-free survival (PFS) in mBC patients by combining baseline radiomics, post-treatment changes (Weeks 8, 16, 24), and demographic features. Radiomics effects were studied across time points separately and through a non-correlated additive framework. Radiomics features were reduced using (a) a regularized (L1-penalized) additive Cox proportional hazards model, and (b) variable selection via best subset selection. Performance, measured using the concordance index (C-index), improved with additional time points. Joint modeling, considering correlations among radiomics effects over time, provided insights into relationships between longitudinal radiomics and survival outcomes.
患有转移性乳腺癌(mBC)的患者在治疗过程中会经历持续性的医学影像检查,因此对病灶进行准确的检测和长时间的监测对临床决策至关重要。从治疗后的数据中预测药物反应对于个性化护理和药物研究至关重要。我们与美国食品药品监督管理局(FDA)和诺华制药公司合作,分析了来自两项大规模III期试验(MONALEESA 3和MONALEESA 7)的连续胸部CT扫描数据。本文有两个目标:(a)开发一种名为注册辅助自动对应(RAMAC)的算法,用于精确追踪纵向CT数据中的病灶;(b)利用RAMAC结构化数据进行生存分析,以预测患者的治疗结果。RAMAC算法采用两阶段流程:三维刚性注册对齐CT图像,基于距离度量的匈牙利算法跟踪病灶对应关系。我们使用结构化数据,结合基线放射组学、治疗后变化(第8周、第16周、第24周)和人口统计学特征,开发了可解释模型来评估mBC患者的无进展生存期(PFS)。放射组学的影响是分别在不同时间点以及通过非相关附加框架进行研究的。使用(a)正则化(L1惩罚)附加Cox比例风险模型和(b)通过最佳子集选择进行变量选择来减少放射学特征。通过增加时间点,以一致性指数(C指数)衡量的性能有所提高。联合建模,考虑到放射学效应在时间上的相关性,提供了纵向放射组学和生存结果之间关系的新见解。
论文及项目相关链接
PDF 20 pages, 13 figures, 2 tables
Summary
本研究针对转移性乳腺癌患者的治疗过程中的医学影像分析,提出了两大目标:一是开发注册辅助自动对应算法(RAMAC),用于精确追踪纵向CT数据中的病灶;二是利用RAMAC结构化数据进行生存分析,预测患者预后。研究通过结合基线放射学特征、治疗后的变化以及人口统计学特征,评估了无进展生存期。通过建模分析,考虑放射学效应在时间上的相关性,揭示了纵向放射学与生存期结果之间的关系。
Key Takeaways
- 研究针对转移性乳腺癌(mBC)患者的治疗过程进行医学影像分析,涉及两大目标:数据结构化和生存分析。
- 开发注册辅助自动对应算法(RAMAC)进行精确病灶追踪。
- RAMAC算法采用两阶段管道处理,包括三维刚性注册对齐CT图像和使用基于距离度量的匈牙利算法追踪病灶对应关系。
- 结合基线放射学特征、治疗后的变化和人口统计学特征,评估无进展生存期(PFS)。
- 放射学特征的影响在不同时间点被单独以及通过一个非相关添加框架进行研究。
- 通过正则化(L1惩罚)的附加Cox比例风险模型和最佳子集选择进行变量选择,以降低放射学特征。
点此查看论文截图

RSRefSeg: Referring Remote Sensing Image Segmentation with Foundation Models
Authors:Keyan Chen, Jiafan Zhang, Chenyang Liu, Zhengxia Zou, Zhenwei Shi
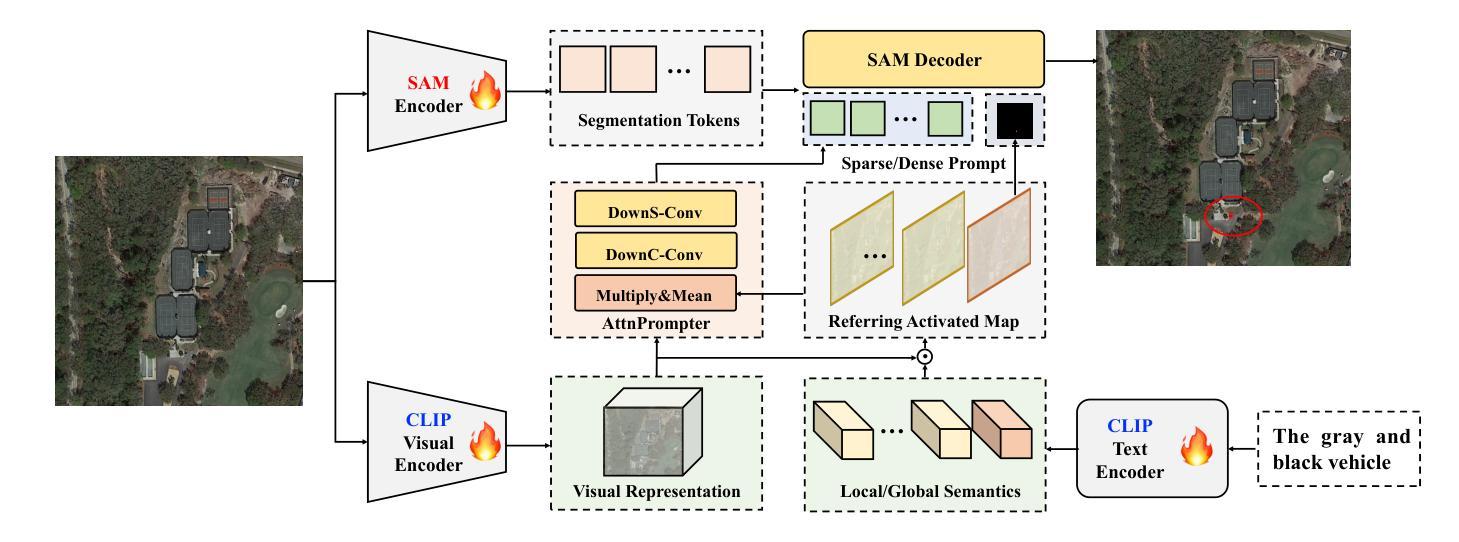
Referring remote sensing image segmentation is crucial for achieving fine-grained visual understanding through free-format textual input, enabling enhanced scene and object extraction in remote sensing applications. Current research primarily utilizes pre-trained language models to encode textual descriptions and align them with visual modalities, thereby facilitating the expression of relevant visual features. However, these approaches often struggle to establish robust alignments between fine-grained semantic concepts, leading to inconsistent representations across textual and visual information. To address these limitations, we introduce a referring remote sensing image segmentation foundational model, RSRefSeg. RSRefSeg leverages CLIP for visual and textual encoding, employing both global and local textual semantics as filters to generate referring-related visual activation features in the latent space. These activated features then serve as input prompts for SAM, which refines the segmentation masks through its robust visual generalization capabilities. Experimental results on the RRSIS-D dataset demonstrate that RSRefSeg outperforms existing methods, underscoring the effectiveness of foundational models in enhancing multimodal task comprehension. The code is available at \url{https://github.com/KyanChen/RSRefSeg}.
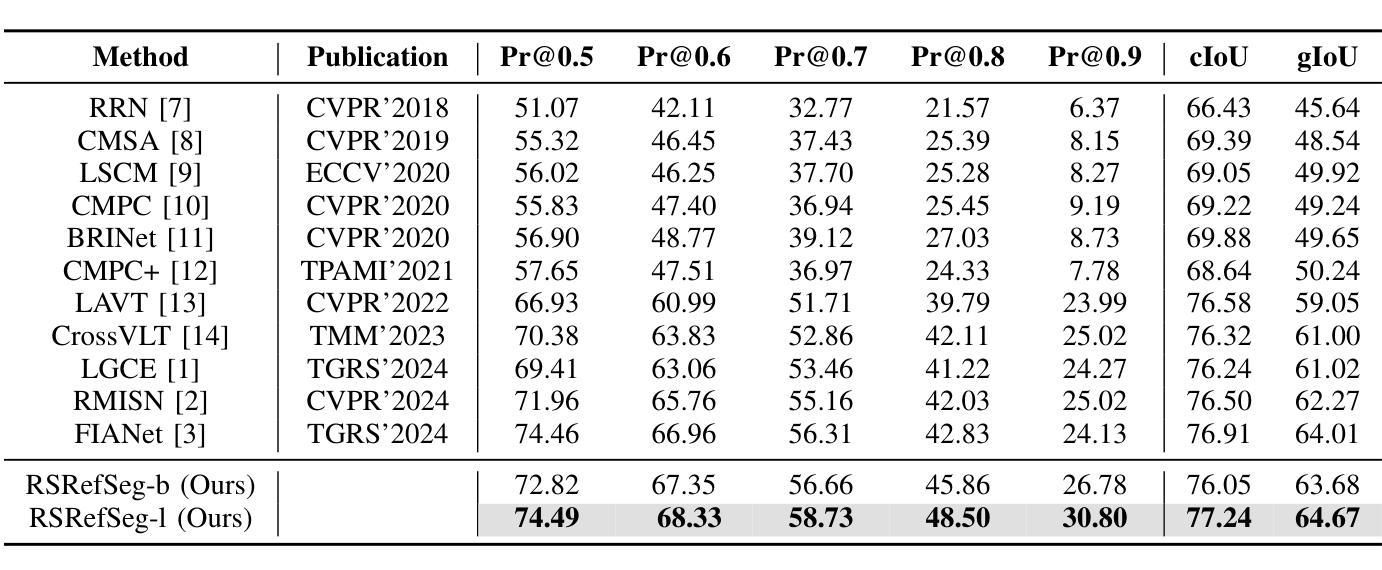
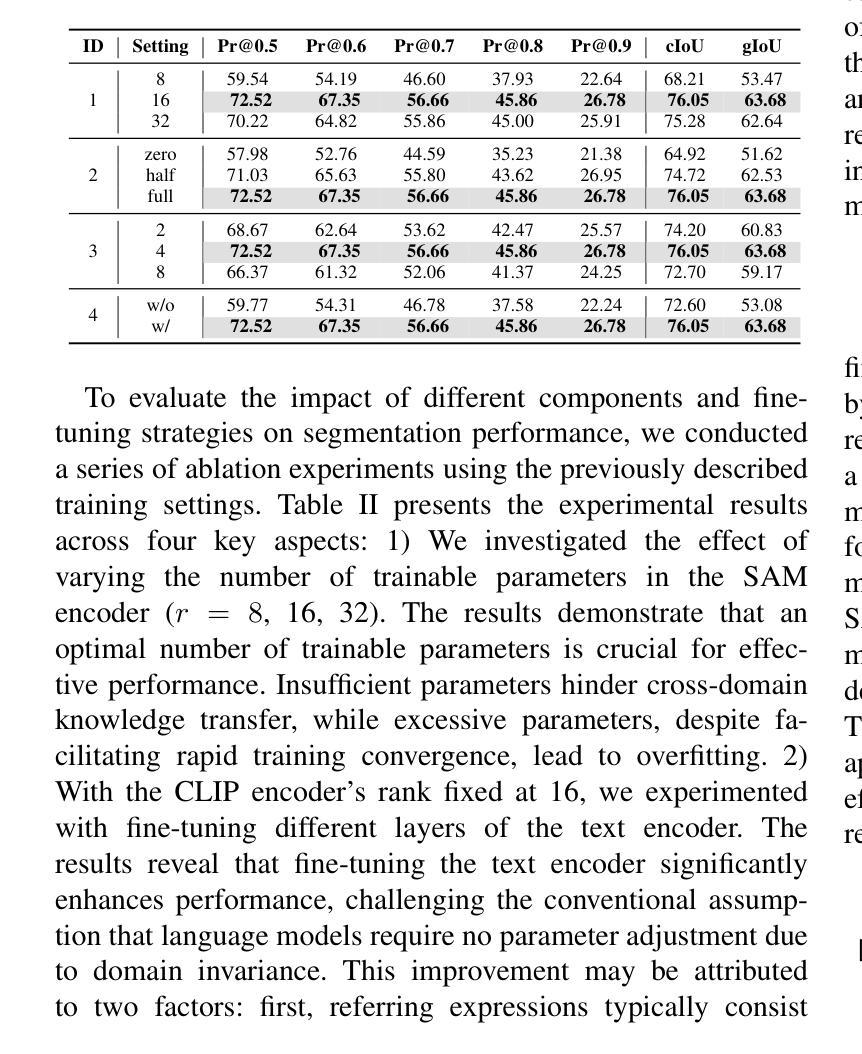
远程遥感图像分割对于通过自由格式的文本输入实现精细的视觉理解至关重要,能够在遥感应用中实现增强的场景和对象提取。目前的研究主要利用预训练的语言模型对文本描述进行编码,并将其与视觉模式对齐,从而促进相关视觉特征的表达。然而,这些方法在建立精细语义概念之间的稳健对齐时往往遇到困难,导致文本和视觉信息之间的表示不一致。为了解决这些局限性,我们引入了一种远程遥感图像分割基础模型RSRefSeg。RSRefSeg利用CLIP进行视觉和文本编码,采用全局和局部文本语义作为过滤器来生成相关的视觉激活特征。这些激活特征随后作为SAM的输入提示,通过其强大的视觉泛化能力来完善分割掩膜。在RRSIS-D数据集上的实验结果表明,RSRefSeg优于现有方法,突显了基础模型在提高多模态任务理解方面的有效性。代码可在https://github.com/KyanChen/RSRefSeg上找到。
论文及项目相关链接
Summary
远程遥感图像分割对于通过自由格式的文本输入实现精细的视觉理解至关重要,有助于增强场景和对象的提取。当前研究主要利用预训练的语言模型对文本描述进行编码,并将其与视觉模式对齐,从而表达相关的视觉特征。然而,这些方法在建立精细语义概念之间的稳健对齐时常常遇到困难,导致跨文本和视觉信息的表示不一致。为解决这些问题,我们引入了遥感图像分割基础模型RSRefSeg。RSRefSeg利用CLIP进行视觉和文本编码,采用全局和局部文本语义作为过滤器,生成与引用相关的视觉激活特征。这些激活特征作为SAM的输入提示,通过其强大的视觉泛化能力细化分割掩膜。在RRSIS-D数据集上的实验结果表明,RSRefSeg优于现有方法,突显了基础模型在增强多模式任务理解方面的有效性。
Key Takeaways
- 远程遥感图像分割对于精细的视觉理解至关重要,有助于场景和对象的增强提取。
- 当前方法主要利用预训练的语言模型进行文本和视觉的编码与对齐。
- 现有方法难以建立精细语义概念之间的稳健对齐,导致跨文本和视觉信息的表示不一致。
- 引入的RSRefSeg模型利用CLIP进行视觉和文本编码。
- RSRefSeg采用全局和局部文本语义作为过滤器,生成与引用相关的视觉激活特征。
- 这些激活特征被用作SAM的输入提示,以细化分割掩膜。
- 在RRSIS-D数据集上的实验显示,RSRefSeg模型优于现有方法,突显了基础模型在增强多模式任务理解方面的有效性。
点此查看论文截图