⚠️ 以下所有内容总结都来自于 大语言模型的能力,如有错误,仅供参考,谨慎使用
🔴 请注意:千万不要用于严肃的学术场景,只能用于论文阅读前的初筛!
💗 如果您觉得我们的项目对您有帮助 ChatPaperFree ,还请您给我们一些鼓励!⭐️ HuggingFace免费体验
2025-02-26 更新
MDN: Mamba-Driven Dualstream Network For Medical Hyperspectral Image Segmentation
Authors:Shijie Lin, Boxiang Yun, Wei Shen, Qingli Li, Anqiang Yang, Yan Wang
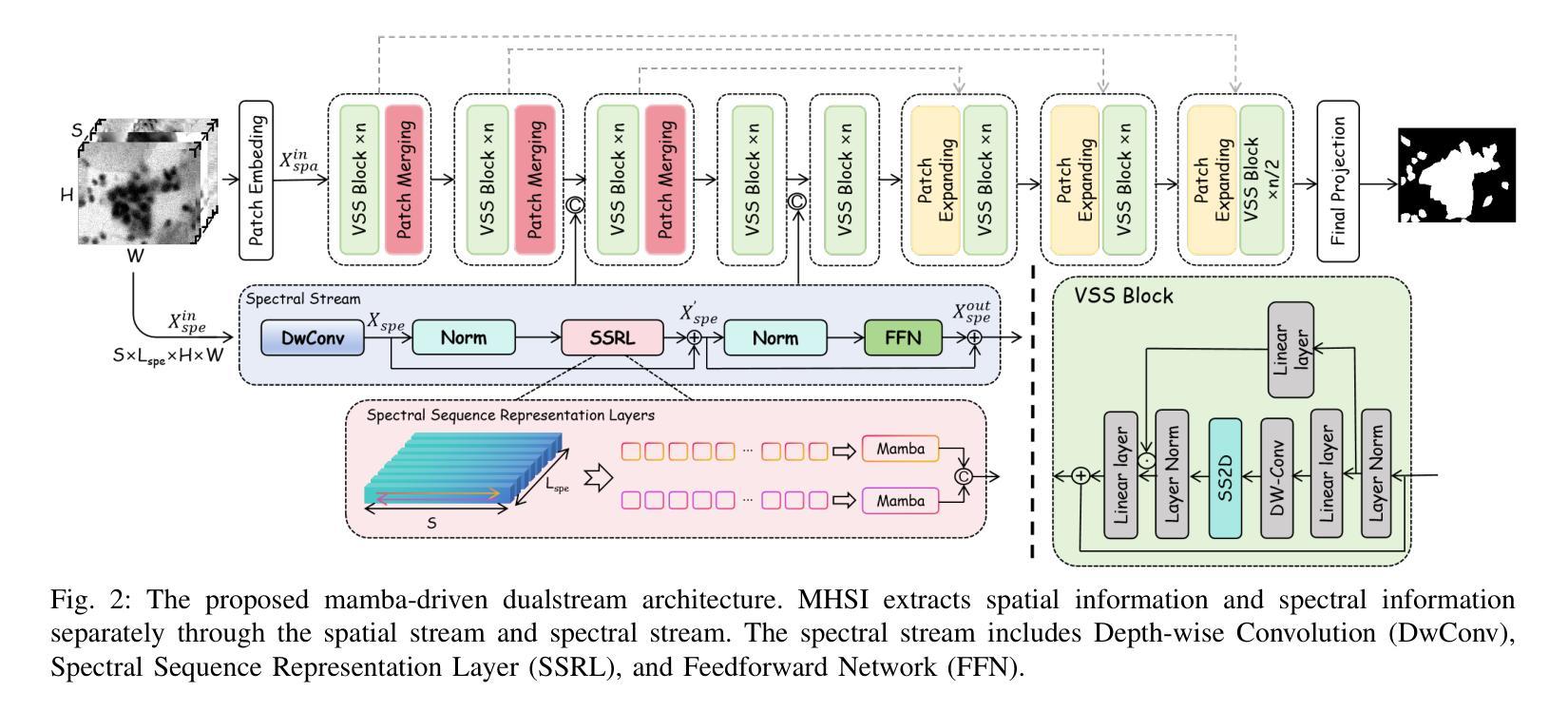
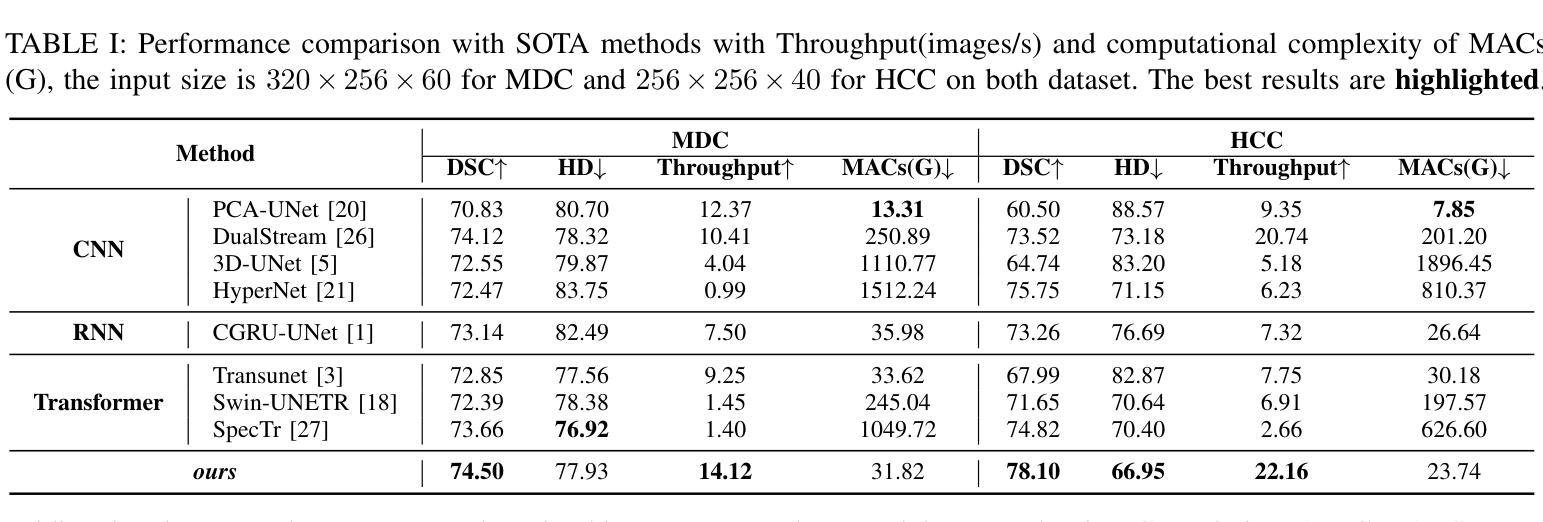
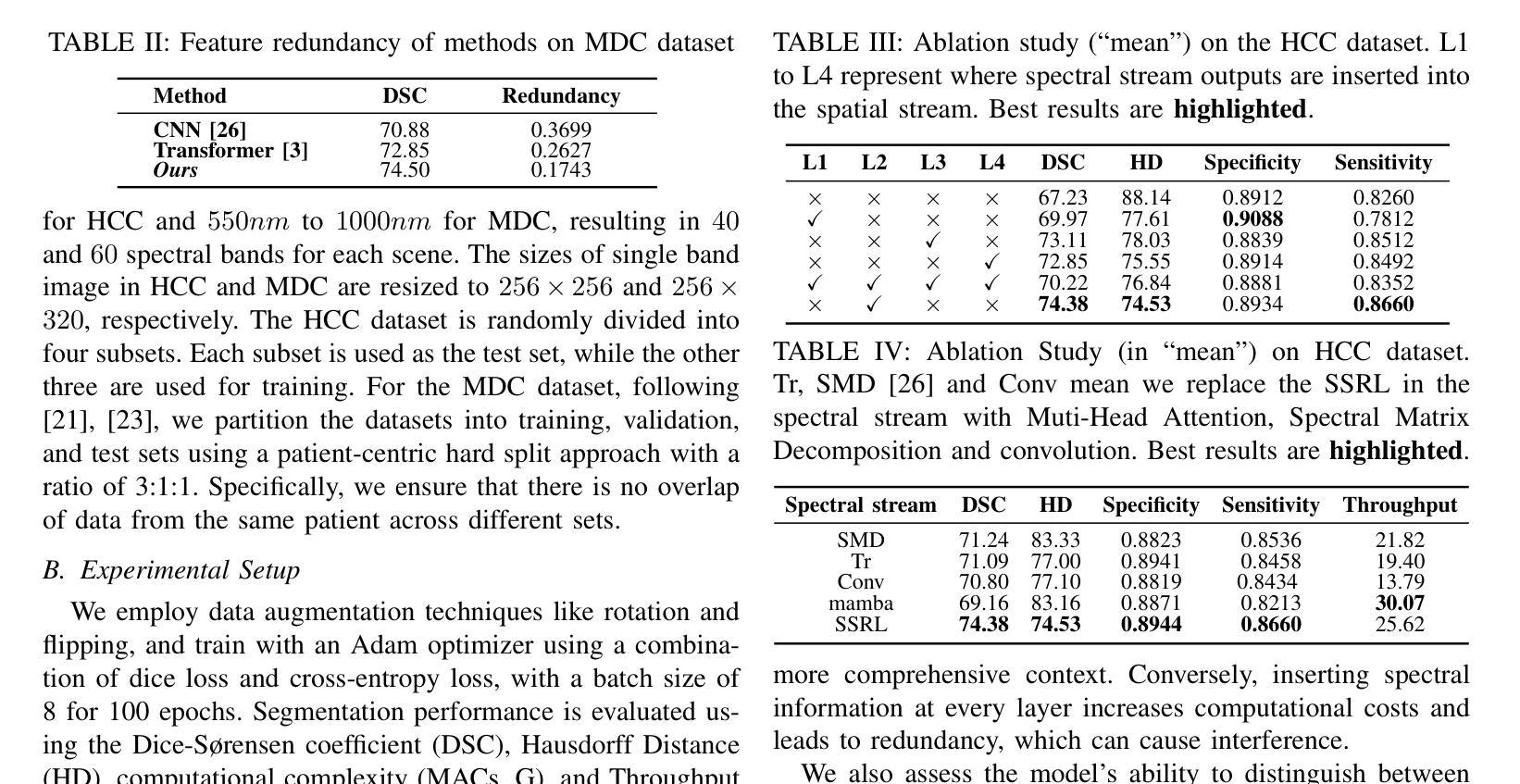
Medical Hyperspectral Imaging (MHSI) offers potential for computational pathology and precision medicine. However, existing CNN and Transformer struggle to balance segmentation accuracy and speed due to high spatial-spectral dimensionality. In this study, we leverage Mamba’s global context modeling to propose a dual-stream architecture for joint spatial-spectral feature extraction. To address the limitation of Mamba’s unidirectional aggregation, we introduce a recurrent spectral sequence representation to capture low-redundancy global spectral features. Experiments on a public Multi-Dimensional Choledoch dataset and a private Cervical Cancer dataset show that our method outperforms state-of-the-art approaches in segmentation accuracy while minimizing resource usage and achieving the fastest inference speed. Our code will be available at https://github.com/DeepMed-Lab-ECNU/MDN.
医学高光谱成像(MHSI)为计算病理学和精准医学提供了潜力。然而,现有的CNN和Transformer由于高空间光谱维度,在平衡分割精度和速度方面存在困难。在这项研究中,我们利用Mamba的全局上下文建模,提出了一种用于联合空间光谱特征提取的双流架构。为了解决Mamba单向聚合的局限性,我们引入递归光谱序列表示,以捕获低冗余全局光谱特征。在公开的Multi-Dimensional Choledoch数据集和私有宫颈癌数据集上的实验表明,我们的方法在分割精度上优于最先进的方法,同时最小化资源使用,实现最快的推理速度。我们的代码将在https://github.com/DeepMed-Lab-ECNU/MDN上提供。
论文及项目相关链接
Summary
医学超光谱成像(MHSI)在计算病理学和精准医学中具有潜力。然而,由于高空间光谱维数,现有CNN和Transformer在分割精度和速度之间难以平衡。本研究利用Mamba的全局上下文建模,提出一种用于联合空间光谱特征提取的双流架构。为解决Mamba单向聚合的局限性,引入循环光谱序列表示法,以捕获低冗余全局光谱特征。在公开的多维胆管数据集和私有宫颈癌数据集上的实验表明,我们的方法在分割精度上优于最新方法,同时最小化资源使用,实现最快的推理速度。
Key Takeaways
- 医学超光谱成像(MHSI)在计算病理学和精准医学中有广泛应用潜力。
- 现有技术如CNN和Transformer在平衡分割精度和速度时面临挑战。
- 研究利用Mamba的全局上下文建模,提出双流架构进行空间光谱特征提取。
- 引入循环光谱序列表示法,解决Mamba单向聚合的局限性。
- 方法在多个数据集上的实验表现优越,包括公开的多维胆管数据集和私有宫颈癌数据集。
- 与现有方法相比,该方法在分割精度上有所提升。
点此查看论文截图

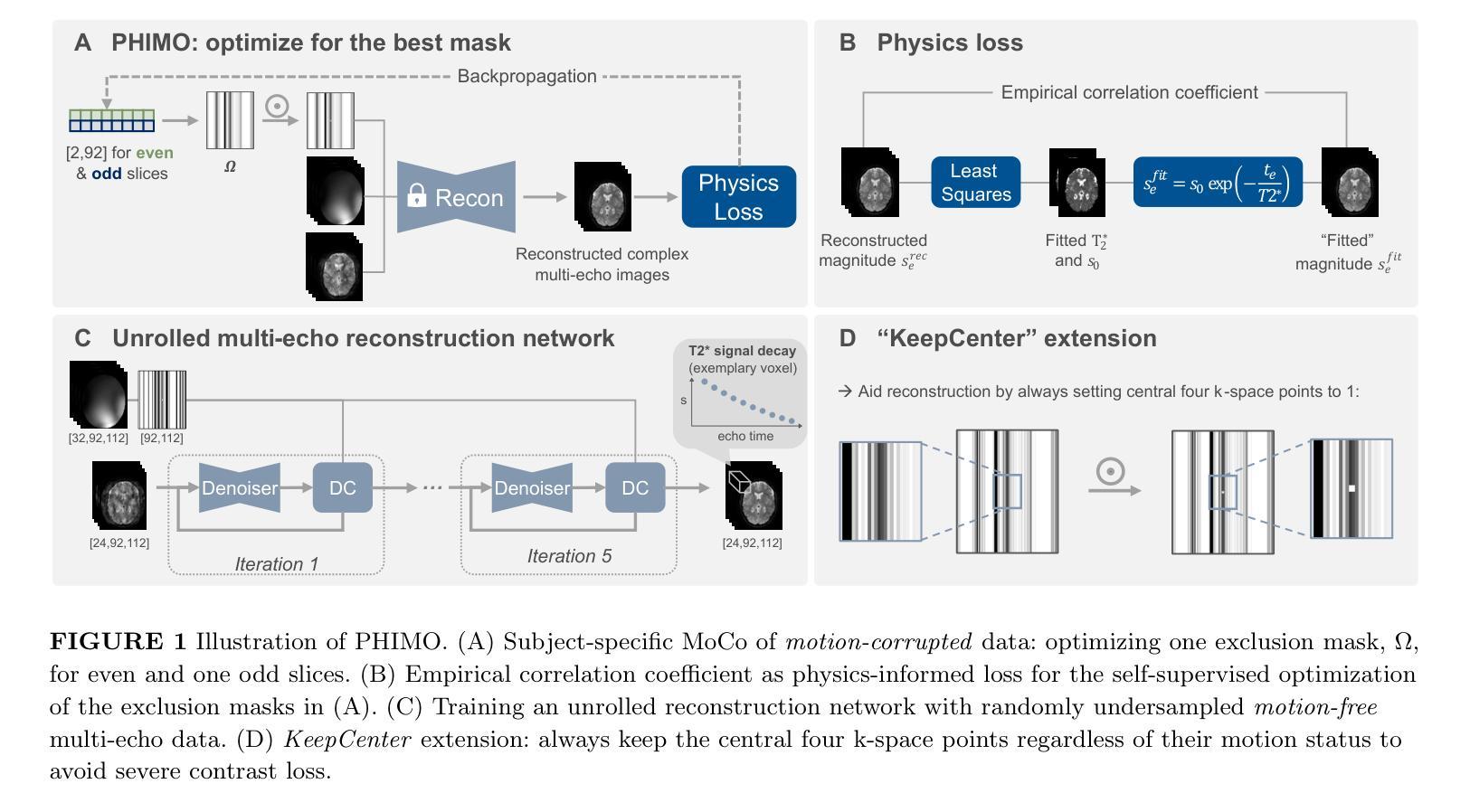
Motion-Robust T2* Quantification from Gradient Echo MRI with Physics-Informed Deep Learning
Authors:Hannah Eichhorn, Veronika Spieker, Kerstin Hammernik, Elisa Saks, Lina Felsner, Kilian Weiss, Christine Preibisch, Julia A. Schnabel
Purpose: T2* quantification from gradient echo magnetic resonance imaging is particularly affected by subject motion due to the high sensitivity to magnetic field inhomogeneities, which are influenced by motion and might cause signal loss. Thus, motion correction is crucial to obtain high-quality T2* maps. Methods: We extend our previously introduced learning-based physics-informed motion correction method, PHIMO, by utilizing acquisition knowledge to enhance the reconstruction performance for challenging motion patterns and increase PHIMO’s robustness to varying strengths of magnetic field inhomogeneities across the brain. We perform comprehensive evaluations regarding motion detection accuracy and image quality for data with simulated and real motion. Results: Our extended version of PHIMO outperforms the learning-based baseline methods both qualitatively and quantitatively with respect to line detection and image quality. Moreover, PHIMO performs on-par with a conventional state-of-the-art motion correction method for T2* quantification from gradient echo MRI, which relies on redundant data acquisition. Conclusion: PHIMO’s competitive motion correction performance, combined with a reduction in acquisition time by over 40% compared to the state-of-the-art method, make it a promising solution for motion-robust T2* quantification in research settings and clinical routine.
目的:从梯度回波磁共振成像获取的T2定量值特别受到受试者运动的影响,因为其对磁场不均匀性具有较高的敏感性,运动会影响磁场不均匀性,并可能导致信号丢失。因此,运动校正对于获得高质量的T2图是至关重要的。方法:我们扩展了我们之前引入的学习型物理信息运动校正方法PHIMO,利用采集知识来提高对复杂运动模式的重建性能,并增强PHIMO对不同强度磁场不均匀性的稳健性。我们对模拟和实际运动的数据进行了全面的运动检测准确性和图像质量评估。结果:我们的PHIMO扩展版本在定性和定量上均优于基于学习的基础方法,特别是在线条检测和图像质量方面。此外,PHIMO与梯度回波MRI的T2定量评估中的常规先进运动校正方法的性能相当,该方法的运动校正依赖于冗余数据采集。结论:PHIMO的竞争性运动校正性能与相比现有技术缩短超过40%的采集时间相结合,使其成为研究和临床实践中稳健的T2定量评估的有前途的解决方案。
论文及项目相关链接
PDF Under Review
Summary
本文旨在解决梯度回波磁共振成像中T2量化受主体运动影响的问题。为提高对运动模式的重建性能并增强PHIMO对磁场不均匀性的稳健性,作者对基于学习的物理信息运动校正方法PHIMO进行了扩展。经过对模拟和实际运动数据的全面评估,发现扩展后的PHIMO在直线检测和图像质量方面均优于基于学习的基础方法,并与传统的T2梯度回波MRI运动校正方法相当。此外,PHIMO将采集时间减少了40%以上,成为研究和临床常规中运动稳健的T2*量化的有前途的解决方案。
Key Takeaways
- T2*量化在梯度回波磁共振成像中易受主体运动影响,导致磁场不均匀性和信号损失。
- PHIMO是一种基于学习和物理信息的运动校正方法,针对磁场不均匀性的不同强度进行了优化。
- 扩展后的PHIMO在模拟和实际运动数据的测试中表现出更高的性能,提高了运动检测准确性和图像质量。
- PHIMO与现有的先进运动校正方法相比,在T2*量化方面具有竞争力。
- PHIMO能够减少超过40%的采集时间,使其成为研究和临床实践中具有吸引力的解决方案。
- PHIMO的应用有助于改善运动稳健的T2*量化在研究和临床常规中的表现。
点此查看论文截图

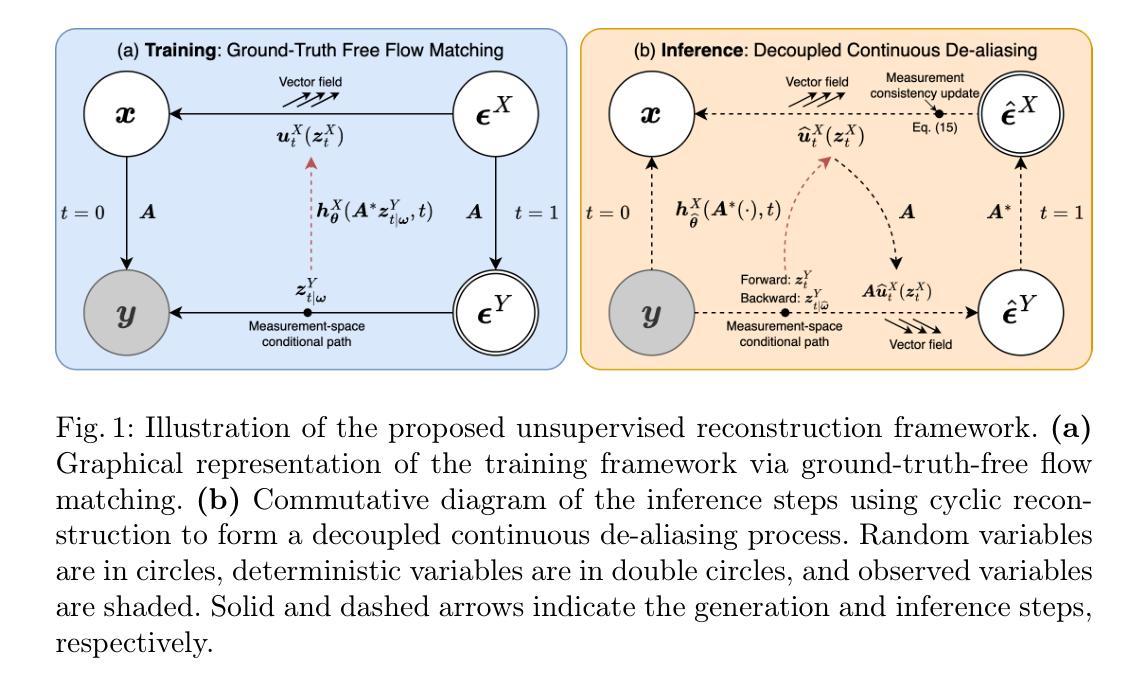
Unsupervised Accelerated MRI Reconstruction via Ground-Truth-Free Flow Matching
Authors:Xinzhe Luo, Yingzhen Li, Chen Qin
Accelerated magnetic resonance imaging involves reconstructing fully sampled images from undersampled k-space measurements. Current state-of-the-art approaches have mainly focused on either end-to-end supervised training inspired by compressed sensing formulations, or posterior sampling methods built on modern generative models. However, their efficacy heavily relies on large datasets of fully sampled images, which may not always be available in practice. To address this issue, we propose an unsupervised MRI reconstruction method based on ground-truth-free flow matching (GTF$^2$M). Particularly, the GTF$^2$M learns a prior denoising process of fully sampled ground-truth images using only undersampled data. Based on that, an efficient cyclic reconstruction algorithm is further proposed to perform forward and backward integration in the dual space of image-space signal and k-space measurement. We compared our method with state-of-the-art learning-based baselines on the fastMRI database of both single-coil knee and multi-coil brain MRIs. The results show that our proposed unsupervised method can significantly outperform existing unsupervised approaches, and achieve performance comparable to most supervised end-to-end and prior learning baselines trained on fully sampled MRI, while offering greater efficiency than the compared generative model-based approaches.
加速磁共振成像涉及从欠采样k空间测量中重建完全采样的图像。目前最前沿的方法主要集中在受压缩感知公式启发的端到端监督训练,或基于现代生成模型的后续采样方法。然而,它们的效力严重依赖于完全采样的图像的大数据集合,这在实践中可能并不总能得到。为了解决这一问题,我们提出了一种基于无真实值流匹配(GTF²M)的无监督MRI重建方法。特别是,GTF²M仅使用欠采样数据学习完全采样的真实值图像的前向去噪过程。在此基础上,进一步提出了一种高效的循环重建算法,在图像空间信号和k空间测量的双重空间中执行正向和反向集成。我们在fastMRI数据库的单线圈膝关节和多线圈脑部MRI上,将我们的方法与基于学习的最新基线进行了比较。结果表明,我们提出的无监督方法可以显著优于现有的无监督方法,并实现与大多数在完全采样的MRI上训练的端到端监督方法和先验学习基线相当的性能,同时提供了比基于生成模型的对比方法更高的效率。
论文及项目相关链接
PDF Accepted by IPMI 2025: 29th International Conference on Information Processing in Medical Imaging
Summary
本文提出了一种基于无真实值流匹配(GTF²M)的无监督MRI重建方法,该方法使用仅了解欠采样数据来模拟真实值的先验去噪过程。通过在该先验基础上进行正向和反向整合,实现了高效的循环重建算法。在fastMRI数据库的膝关节单线圈和大脑多线圈MRI测试中,该方法显著优于现有无监督方法,性能与大多数基于完全采样MRI训练的端到端和先验学习基线相当,同时比基于生成模型的对比方法更高效。
Key Takeaways
- 论文提出了一种基于无真实值流匹配(GTF²M)的无监督MRI重建方法。
- 该方法通过模拟真实值的先验去噪过程来学习先验知识,仅使用欠采样数据。
- 通过结合循环重建算法实现了高效的前向和后向整合在图像空间信号和k空间测量之间的双重空间。
- 在fastMRI数据库测试中,该方法显著优于现有无监督方法。
- 该方法的性能与大多数基于完全采样MRI训练的端到端和先验学习基线相当。
- 与对比的生成模型方法相比,该方法更高效。
- 此方法对于数据集的依赖程度较低,尤其在没有大量完全采样图像的情况下具有显著优势。
点此查看论文截图

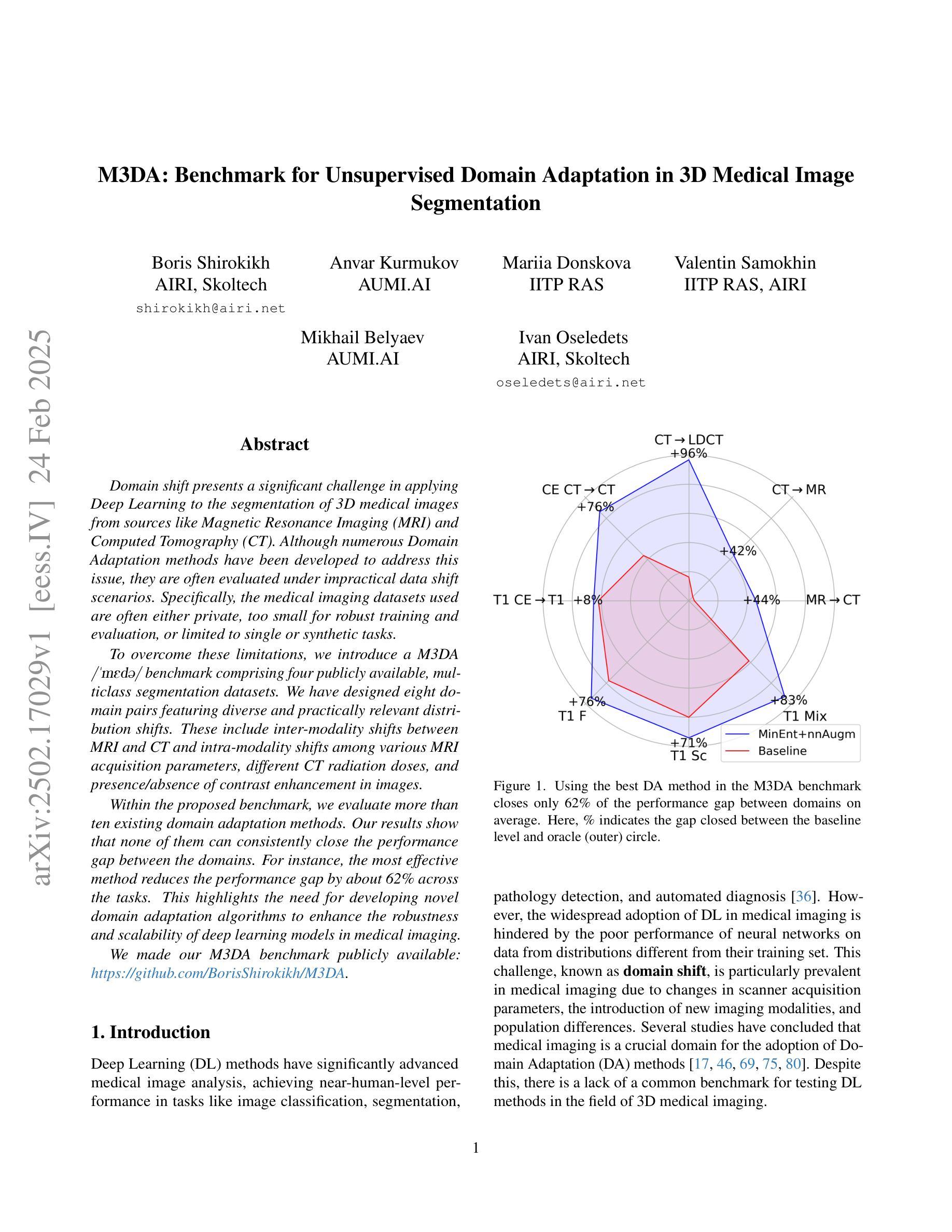
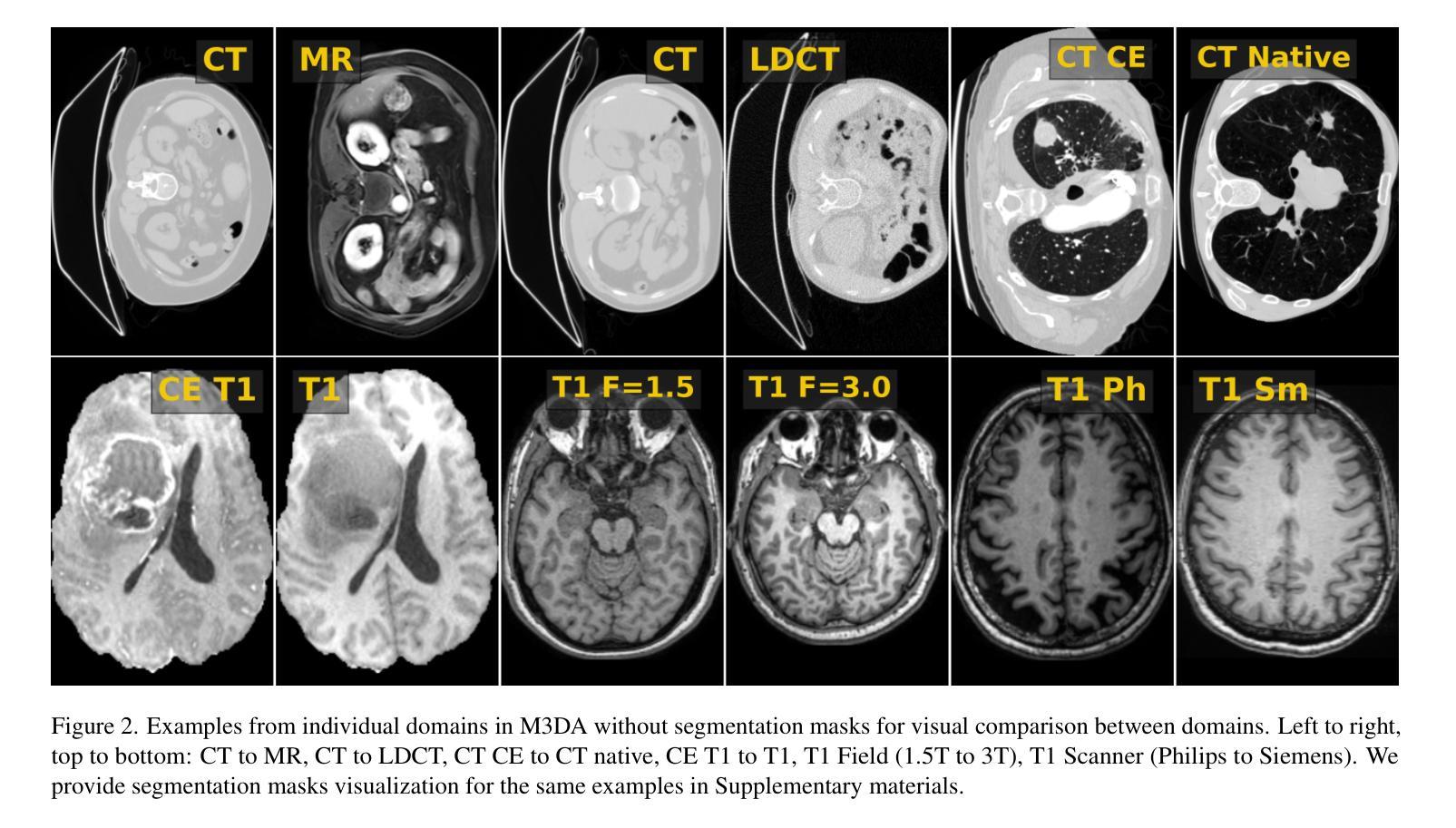
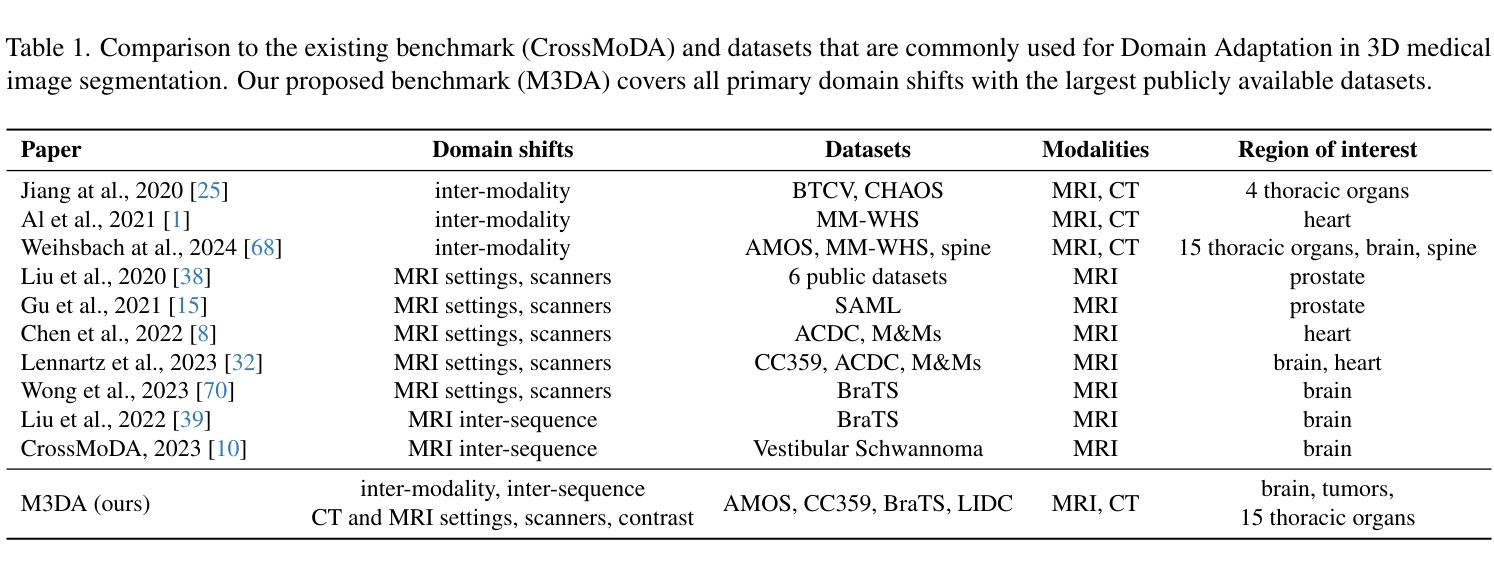
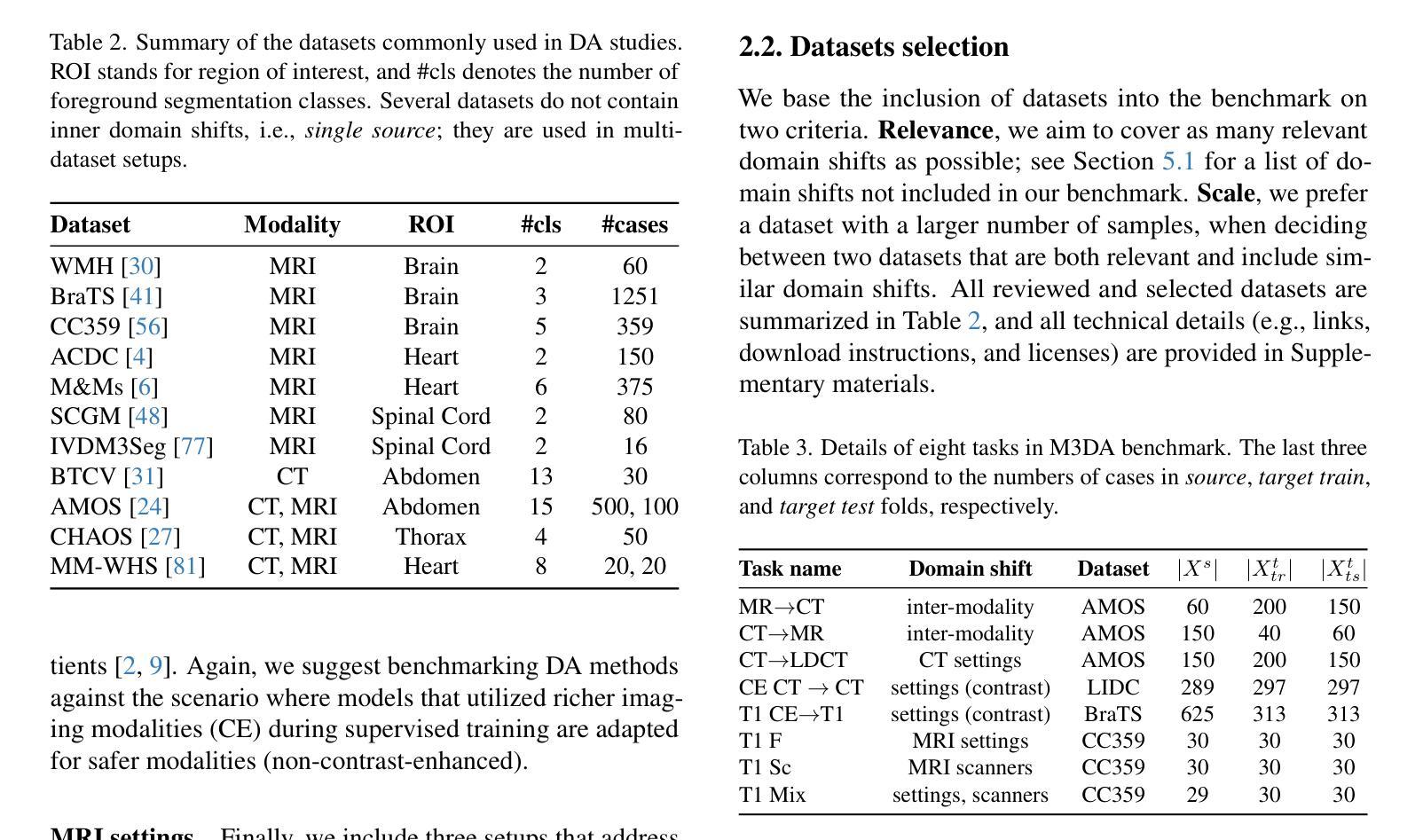
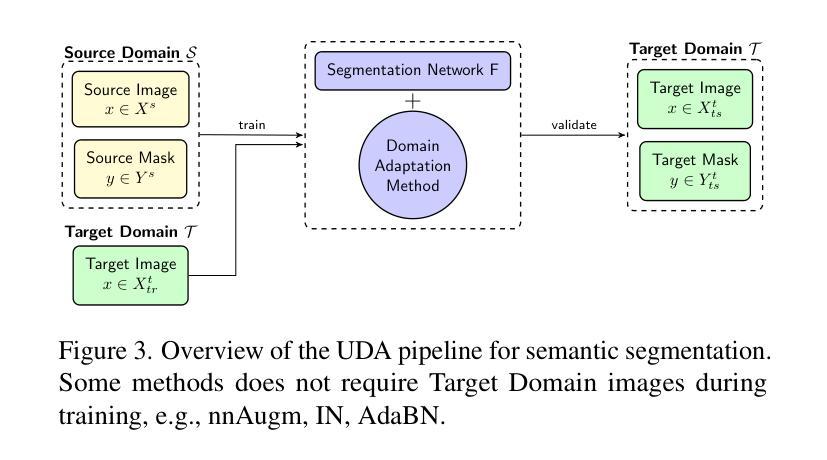
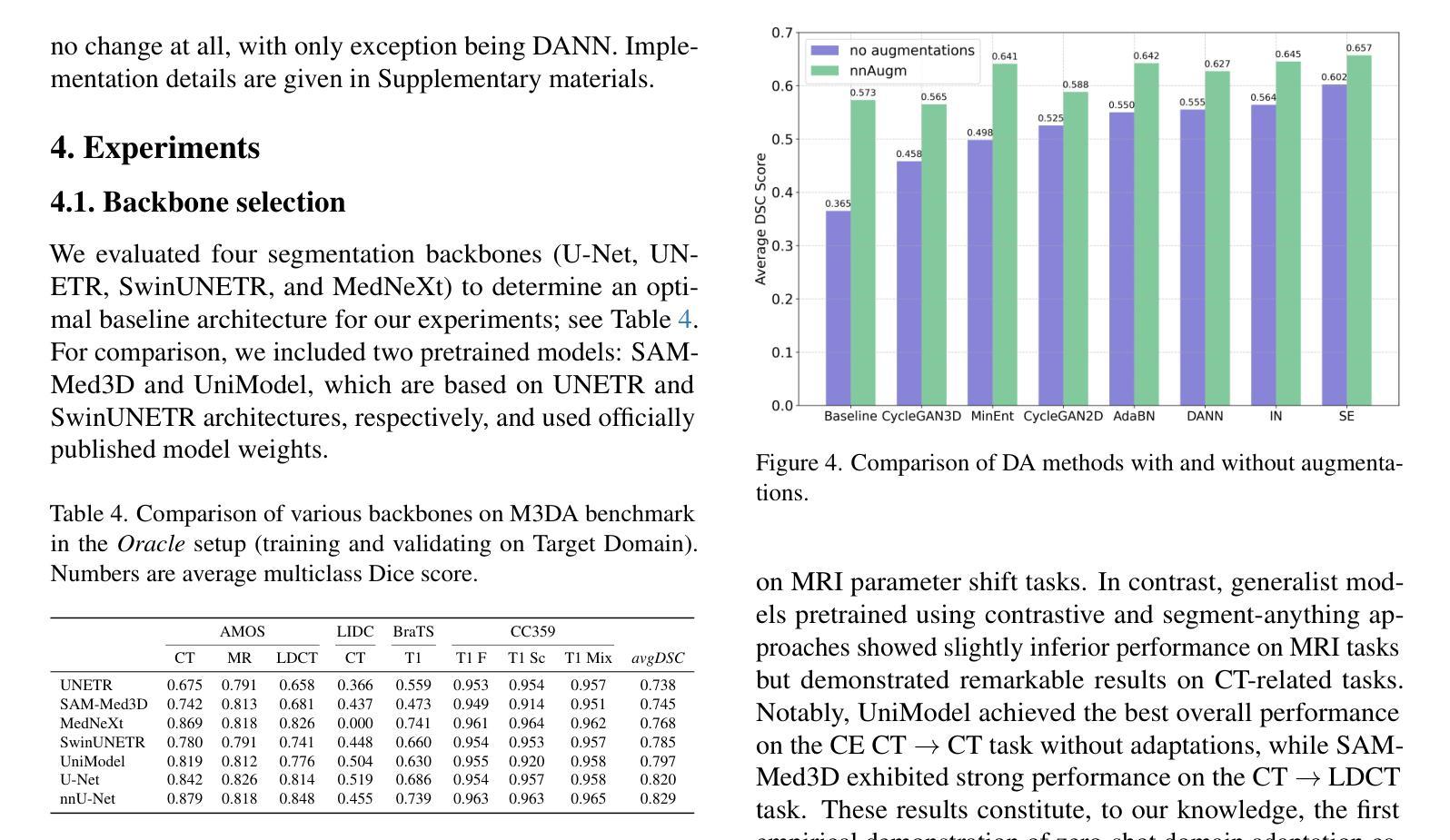
M3DA: Benchmark for Unsupervised Domain Adaptation in 3D Medical Image Segmentation
Authors:Boris Shirokikh, Anvar Kurmukov, Mariia Donskova, Valentin Samokhin, Mikhail Belyaev, Ivan Oseledets
Domain shift presents a significant challenge in applying Deep Learning to the segmentation of 3D medical images from sources like Magnetic Resonance Imaging (MRI) and Computed Tomography (CT). Although numerous Domain Adaptation methods have been developed to address this issue, they are often evaluated under impractical data shift scenarios. Specifically, the medical imaging datasets used are often either private, too small for robust training and evaluation, or limited to single or synthetic tasks. To overcome these limitations, we introduce a M3DA /“mEd@/ benchmark comprising four publicly available, multiclass segmentation datasets. We have designed eight domain pairs featuring diverse and practically relevant distribution shifts. These include inter-modality shifts between MRI and CT and intra-modality shifts among various MRI acquisition parameters, different CT radiation doses, and presence/absence of contrast enhancement in images. Within the proposed benchmark, we evaluate more than ten existing domain adaptation methods. Our results show that none of them can consistently close the performance gap between the domains. For instance, the most effective method reduces the performance gap by about 62% across the tasks. This highlights the need for developing novel domain adaptation algorithms to enhance the robustness and scalability of deep learning models in medical imaging. We made our M3DA benchmark publicly available: https://github.com/BorisShirokikh/M3DA.
领域偏移(Domain Shift)在将深度学习应用于从磁共振成像(MRI)和计算机断层扫描(CT)等来源的3D医学图像分割中带来了重大挑战。尽管已经开发了许多域自适应(Domain Adaptation)方法来应对这个问题,但它们通常是在不切实际的数据偏移场景下进行评估的。具体来说,医学成像数据集通常是私有的,对于稳健训练和评估而言规模太小,或者仅限于单一或合成任务。为了克服这些局限性,我们引入了M3DA基准测试集,其中包含四个公开可用的多类别分割数据集。我们设计了八个具有多样性和实际相关分布偏移的域对。这些包括MRI和CT之间的模态间偏移以及不同MRI采集参数、不同CT辐射剂量和图像中是否存在对比增强之间的模态内偏移。在提出的基准测试集中,我们对超过十种现有的域自适应方法进行了评估。我们的结果表明,没有任何方法可以始终如一地缩小域之间的性能差距。例如,最有效的方法在各项任务中将性能差距减少了约62%。这强调了开发新型域自适应算法的需要,以提高医学成像中深度学习模型的稳健性和可扩展性。我们的M3DA基准测试集已公开可用:https://github.com/BorisShirokikh/M3DA。
论文及项目相关链接
PDF 17 pages,7 figures,11 tables
Summary
本文介绍了在将深度学习应用于三维医学图像分割(如磁共振成像和计算机断层扫描)时面临的挑战,即领域偏移问题。尽管已经开发了许多领域适应方法来应对这一问题,但它们在现实的数据偏移场景中并不实用。为解决此问题,本文提出了一个包含四个公开可用、多类别分割数据集的M3DA基准测试平台,设计了涵盖多种实际相关的分布偏移的八个领域对。评估结果显示,现有的领域适应方法无法一致缩小领域间的性能差距,因此需要开发新的领域适应算法来提高深度学习模型在医学成像中的鲁棒性和可扩展性。
Key Takeaways
- 领域偏移是深度学习在医学图像分割中面临的挑战。
- 现有领域适应方法在实践中并不实用,因为它们通常在私有数据集上评估,数据量小且限于单一任务。
- 提出了一种新的M3DA基准测试平台,包含四个公开的多类别分割数据集。
- 设计了八个具有实际意义的领域对,涵盖多种分布偏移,包括MRI和CT之间的模态偏移以及不同MRI采集参数、CT辐射剂量和有/无图像对比增强的模态内偏移。
- 对超过十种现有的领域适应方法进行了评估,但结果并不理想,没有一种方法可以始终如一地缩小领域间的性能差距。
- 最有效的方法将性能差距减少了约62%,但仍需开发新的领域适应算法来提高模型的鲁棒性和可扩展性。
点此查看论文截图
MAD-AD: Masked Diffusion for Unsupervised Brain Anomaly Detection
Authors:Farzad Beizaee, Gregory Lodygensky, Christian Desrosiers, Jose Dolz
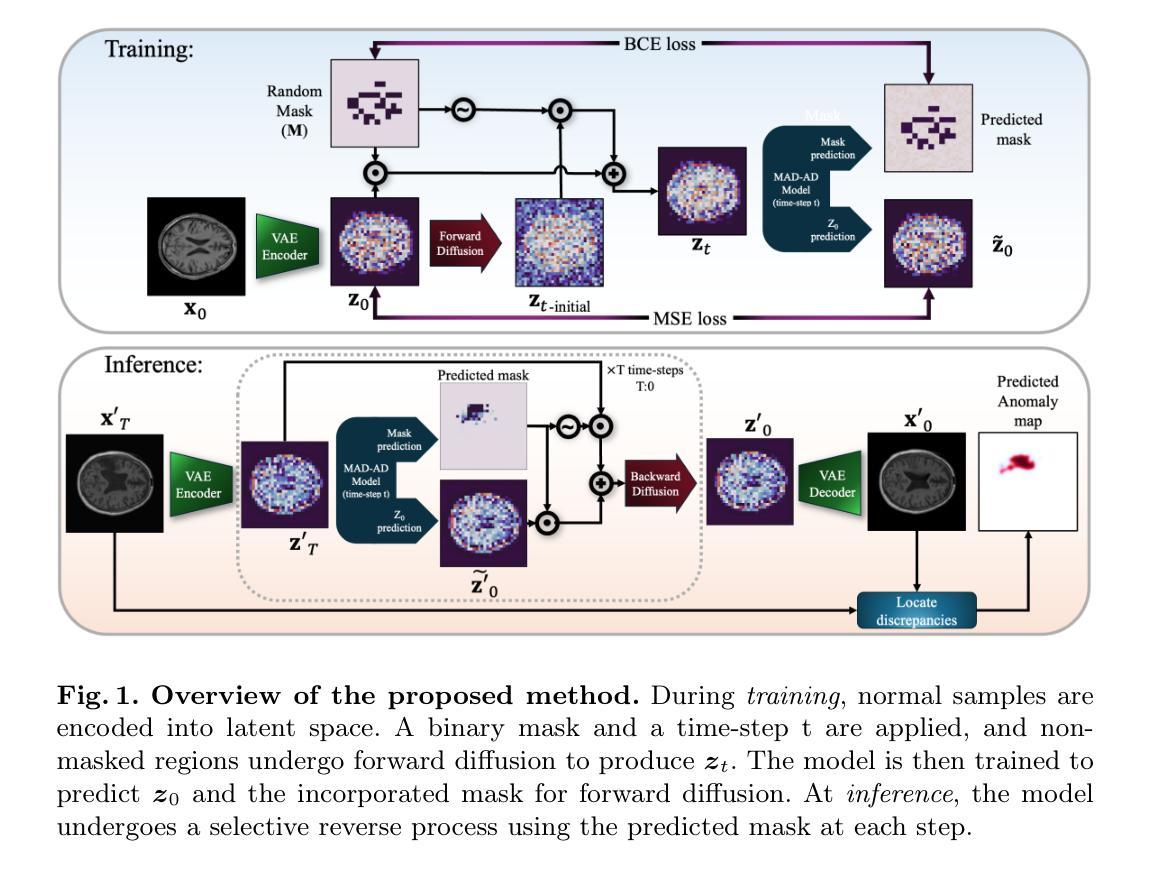
Unsupervised anomaly detection in brain images is crucial for identifying injuries and pathologies without access to labels. However, the accurate localization of anomalies in medical images remains challenging due to the inherent complexity and variability of brain structures and the scarcity of annotated abnormal data. To address this challenge, we propose a novel approach that incorporates masking within diffusion models, leveraging their generative capabilities to learn robust representations of normal brain anatomy. During training, our model processes only normal brain MRI scans and performs a forward diffusion process in the latent space that adds noise to the features of randomly-selected patches. Following a dual objective, the model learns to identify which patches are noisy and recover their original features. This strategy ensures that the model captures intricate patterns of normal brain structures while isolating potential anomalies as noise in the latent space. At inference, the model identifies noisy patches corresponding to anomalies and generates a normal counterpart for these patches by applying a reverse diffusion process. Our method surpasses existing unsupervised anomaly detection techniques, demonstrating superior performance in generating accurate normal counterparts and localizing anomalies. The code is available at hhttps://github.com/farzad-bz/MAD-AD.
在无需标签的情况下,对脑图像进行无监督的异常检测对于识别损伤和病理至关重要。然而,由于脑结构固有的复杂性和可变性以及标注异常数据的稀缺性,准确地在医学图像中定位异常仍然是一个挑战。为了应对这一挑战,我们提出了一种结合扩散模型中遮罩技术的新方法,利用其生成能力学习正常脑解剖结构的稳健表示。在训练过程中,我们的模型仅处理正常的脑部MRI扫描,并在潜在空间执行正向扩散过程,向随机选择的补丁的特征添加噪声。遵循双重目标,模型学习识别哪些补丁是嘈杂的并恢复其原始特征。此策略确保模型捕捉正常脑结构的精细模式,同时将潜在的异常孤立为潜在空间中的噪声。在推理阶段,模型会识别对应于异常的嘈杂补丁,并通过反向扩散过程生成这些补丁的正常对应物。我们的方法超越了现有的无监督异常检测技术,在生成准确的正常对应物和定位异常方面表现出卓越的性能。代码可通过以下链接获取:https://github.com/farzad-bz/MAD-AD。
论文及项目相关链接
Summary
医学图像中的无监督异常检测对于在没有标签的情况下识别损伤和病理至关重要。针对医学图像中异常准确定位的挑战,我们提出了一种结合扩散模型内掩蔽的新方法,利用生成能力学习正常脑结构的有力表征。我们的模型仅处理正常的脑部MRI扫描,在潜在空间中进行正向扩散过程,对随机选择的斑块添加噪声。模型学习识别哪些斑块是嘈杂的并恢复其原始特征,确保模型捕捉正常脑结构的复杂模式,同时将潜在的异常孤立为潜在空间中的噪声。在推理时,模型识别与异常相对应的嘈杂斑块,并通过反向扩散过程为这些斑块生成正常对应物。我们的方法在生成准确的正常对应物和定位异常方面超越了现有的无监督异常检测技术。
Key Takeaways
- 无监督异常检测在医学图像中至关重要,特别是在缺乏标签的情况下。
- 提出了一种结合扩散模型内掩蔽的新方法,用于异常检测。
- 模型通过正向扩散过程学习正常脑结构的表征。
- 模型能够识别嘈杂的斑块并恢复其原始特征。
- 该方法能够在潜在空间中区分正常结构与异常。
- 通过反向扩散过程,模型能生成与异常斑块对应的正常对应物。
- 该方法超越了现有的无监督异常检测技术,表现出优越的性能。
点此查看论文截图

DiffKAN-Inpainting: KAN-based Diffusion model for brain tumor inpainting
Authors:Tianli Tao, Ziyang Wang, Han Zhang, Theodoros N. Arvanitis, Le Zhang
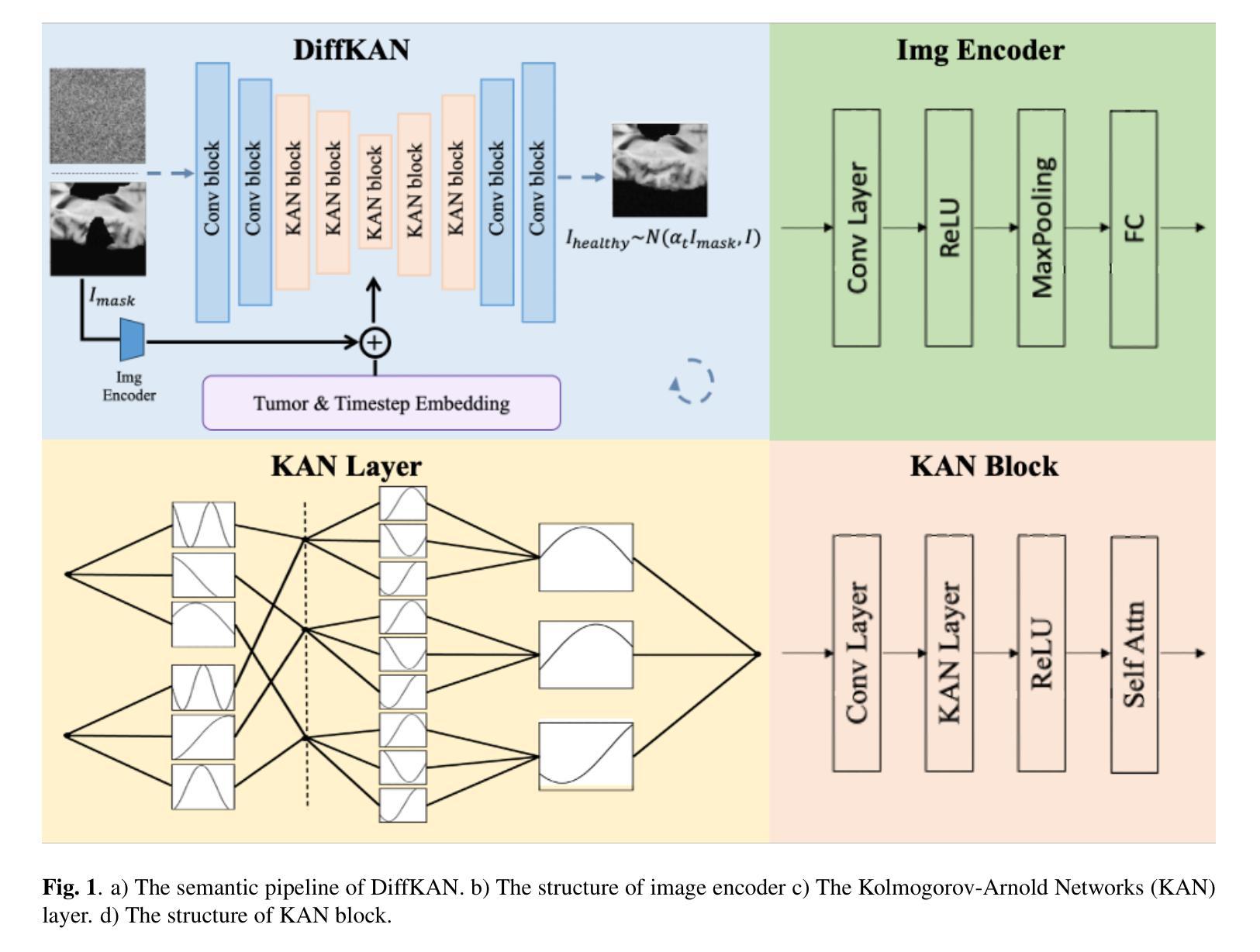
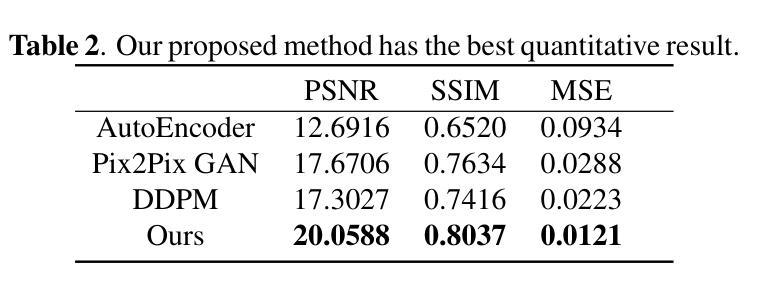
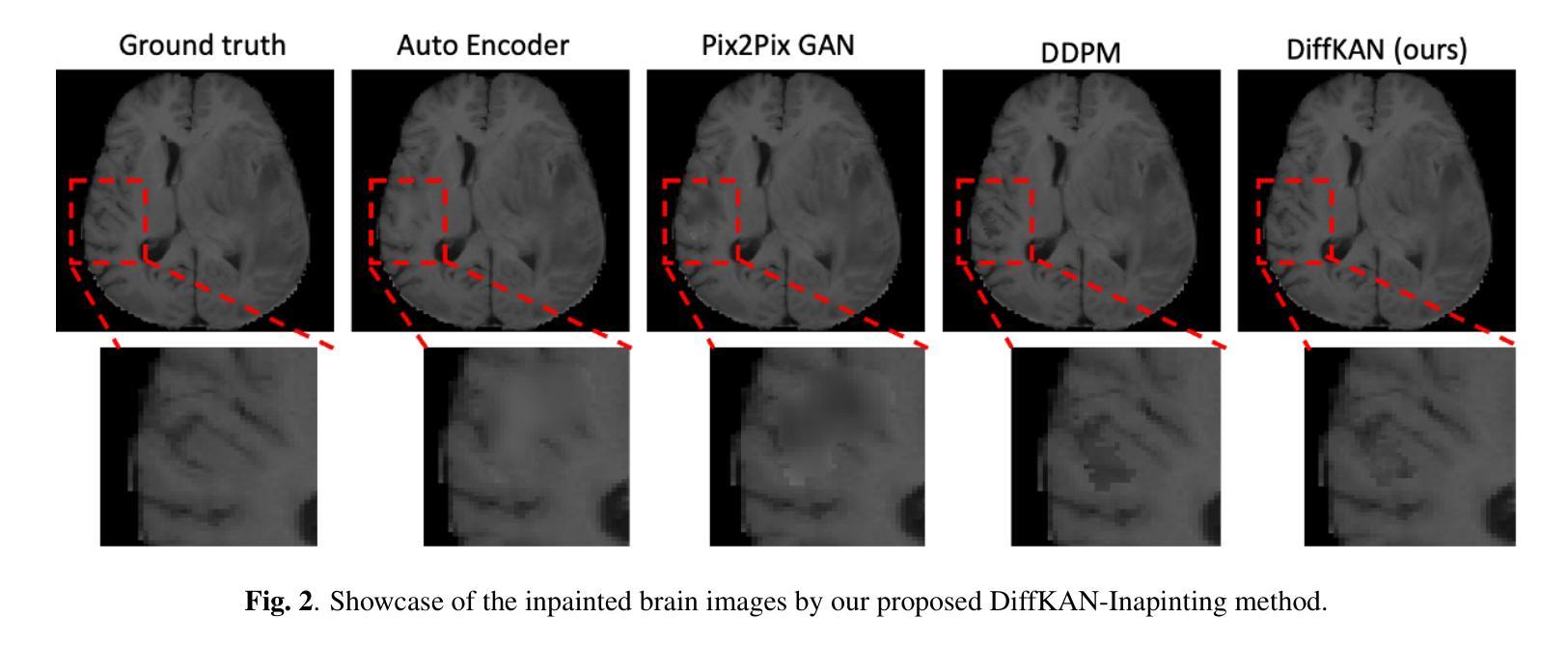
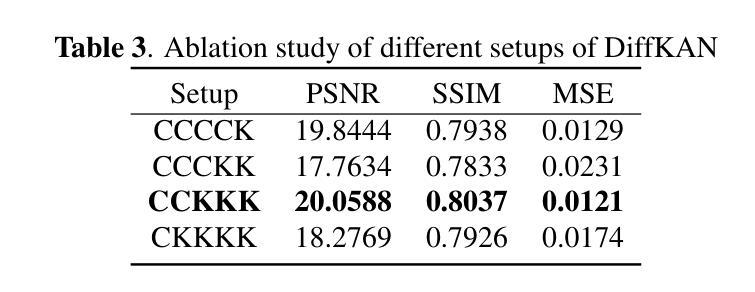
Brain tumors delay the standard preprocessing workflow for further examination. Brain inpainting offers a viable, although difficult, solution for tumor tissue processing, which is necessary to improve the precision of the diagnosis and treatment. Most conventional U-Net-based generative models, however, often face challenges in capturing the complex, nonlinear latent representations inherent in brain imaging. In order to accomplish high-quality healthy brain tissue reconstruction, this work proposes DiffKAN-Inpainting, an innovative method that blends diffusion models with the Kolmogorov-Arnold Networks architecture. During the denoising process, we introduce the RePaint method and tumor information to generate images with a higher fidelity and smoother margin. Both qualitative and quantitative results demonstrate that as compared to the state-of-the-art methods, our proposed DiffKAN-Inpainting inpaints more detailed and realistic reconstructions on the BraTS dataset. The knowledge gained from ablation study provide insights for future research to balance performance with computing cost.
脑肿瘤会延迟进一步的检查标准预处理工作流程。脑填充(Brain inpainting)为肿瘤组织处理提供了一种可行虽然困难的解决方案,这对于提高诊断和治疗精度是必要的。然而,大多数基于U-Net的传统生成模型在捕获脑成像中固有的复杂非线性潜在表示方面面临挑战。为了完成高质量的健康脑组织重建,这项工作提出了DiffKAN-Inpainting这一创新方法,它将扩散模型与Kolmogorov-Arnold网络架构相结合。在降噪过程中,我们引入了RePaint方法和肿瘤信息,以生成具有更高保真度和更平滑边界的图像。定性和定量结果均表明,与最新方法相比,我们提出的DiffKAN-Inpainting在BraTS数据集上进行了更详细和更现实的重建。从消融研究中获得的知识为未来研究提供了在性能与计算成本之间取得平衡的见解。
论文及项目相关链接
Summary
本文提出一种结合扩散模型和Kolmogorov-Arnold网络架构的DiffKAN-Inpainting方法,用于高质量重建健康脑组织。该方法在去除噪声过程中引入RePaint方法和肿瘤信息,生成更逼真、细节更丰富的图像。相较于现有方法,该方法在BraTS数据集上有更好的表现。
Key Takeaways
- Brain tumors影响预处理工作流程,需要进一步处理以改善诊断和治疗的准确性。
- Brain inpainting为解决这个问题提供了一种可行的解决方案。
- 传统的U-Net-based generative模型在捕获脑成像中的复杂非线性潜在表示时面临挑战。
- DiffKAN-Inpainting方法结合了扩散模型和Kolmogorov-Arnold网络架构,旨在实现高质量的健康脑组织重建。
- RePaint方法和肿瘤信息被引入到去噪过程中,生成了更高保真度和更平滑边缘的图像。
- 与最新方法相比,DiffKAN-Inpainting在BraTS数据集上表现出了更好的重建效果和细节表现。
点此查看论文截图

AeroReformer: Aerial Referring Transformer for UAV-based Referring Image Segmentation
Authors:Rui Li
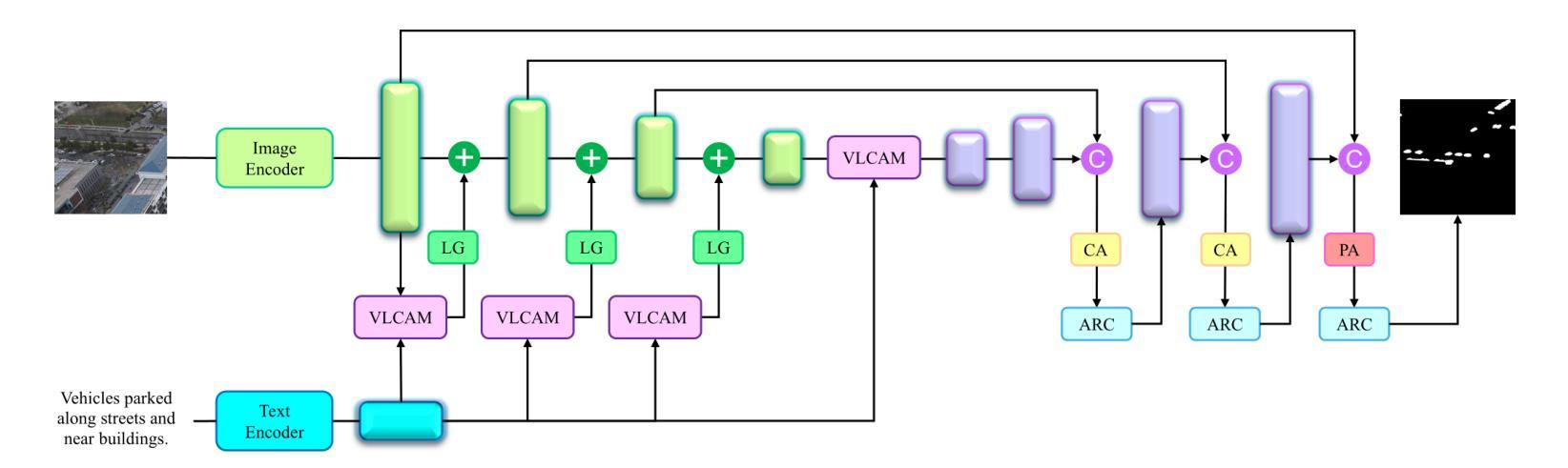
As a novel and challenging task, referring segmentation combines computer vision and natural language processing to localize and segment objects based on textual descriptions. While referring image segmentation (RIS) has been extensively studied in natural images, little attention has been given to aerial imagery, particularly from unmanned aerial vehicles (UAVs). The unique challenges of UAV imagery, including complex spatial scales, occlusions, and varying object orientations, render existing RIS approaches ineffective. A key limitation has been the lack of UAV-specific datasets, as manually annotating pixel-level masks and generating textual descriptions is labour-intensive and time-consuming. To address this gap, we design an automatic labelling pipeline that leverages pre-existing UAV segmentation datasets and Multimodal Large Language Models (MLLM) for generating textual descriptions. Furthermore, we propose Aerial Referring Transformer (AeroReformer), a novel framework for UAV referring image segmentation (UAV-RIS), featuring a Vision-Language Cross-Attention Module (VLCAM) for effective cross-modal understanding and a Rotation-Aware Multi-Scale Fusion (RAMSF) decoder to enhance segmentation accuracy in aerial scenes. Extensive experiments on two newly developed datasets demonstrate the superiority of AeroReformer over existing methods, establishing a new benchmark for UAV-RIS. The datasets and code will be publicly available at: https://github.com/lironui/AeroReformer.
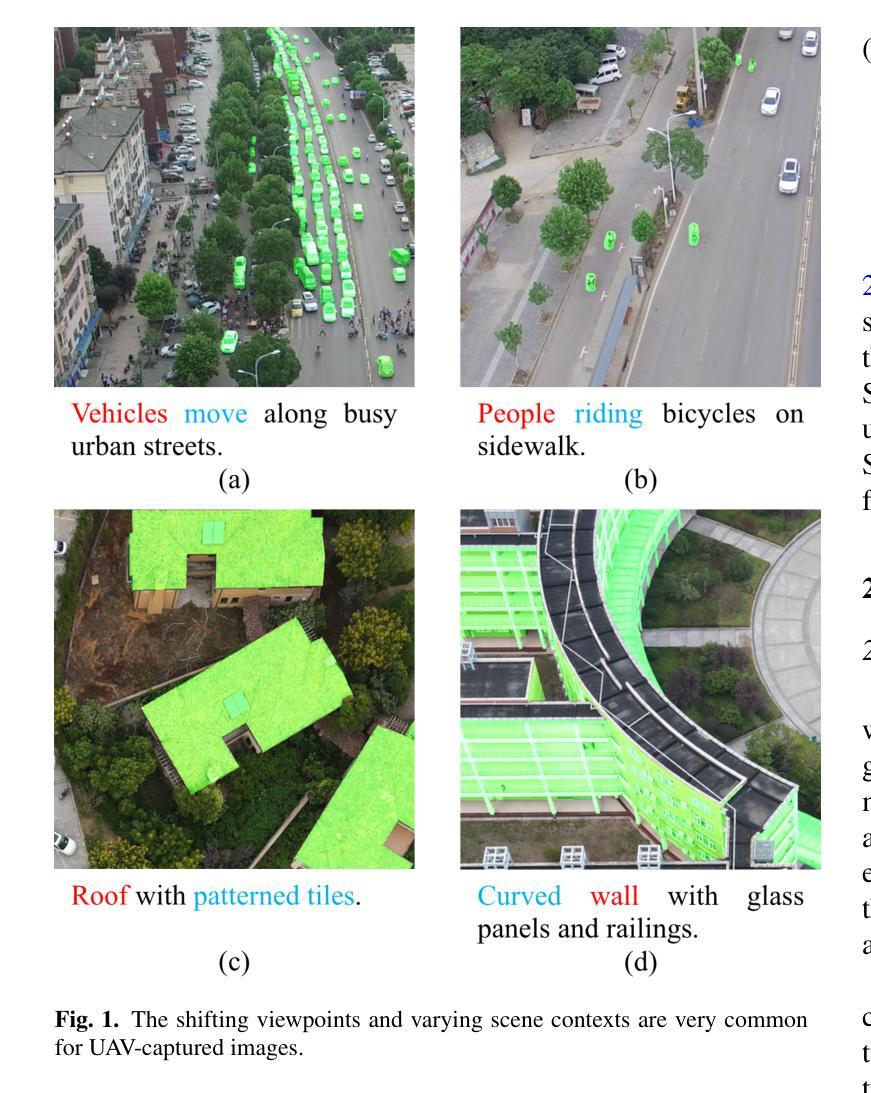
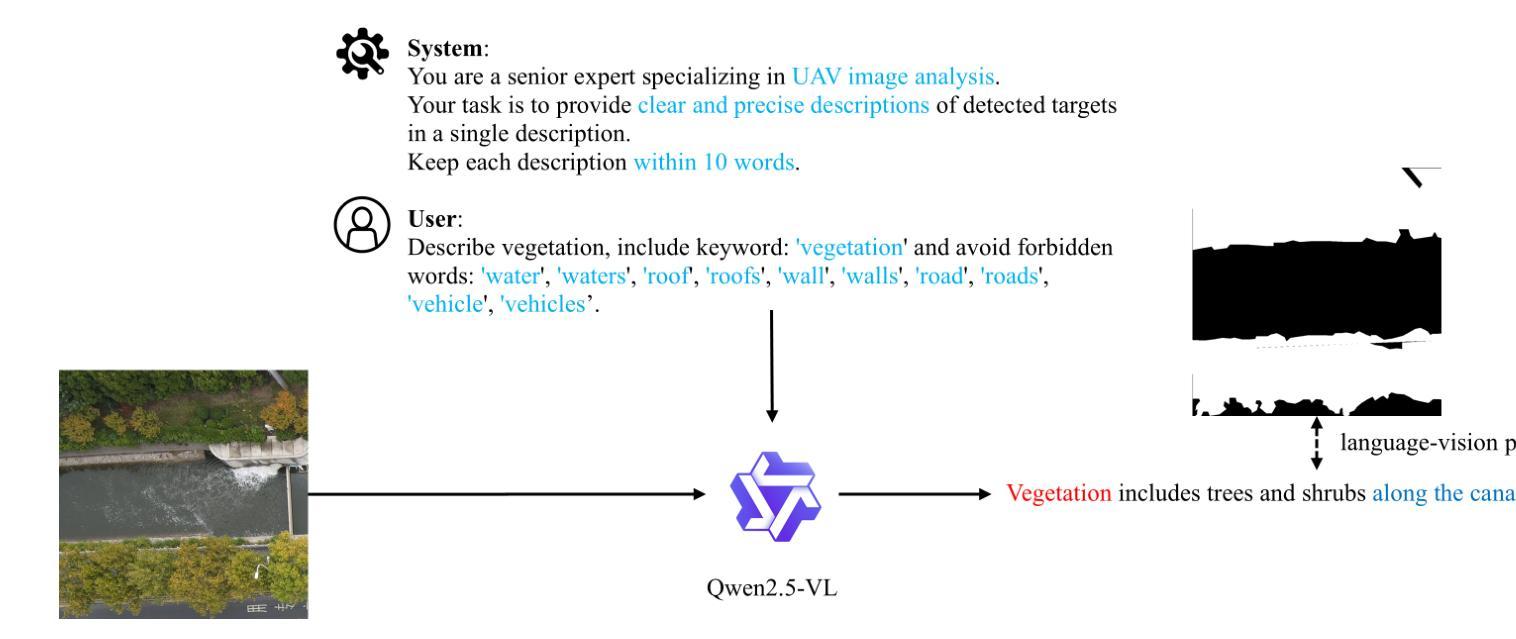
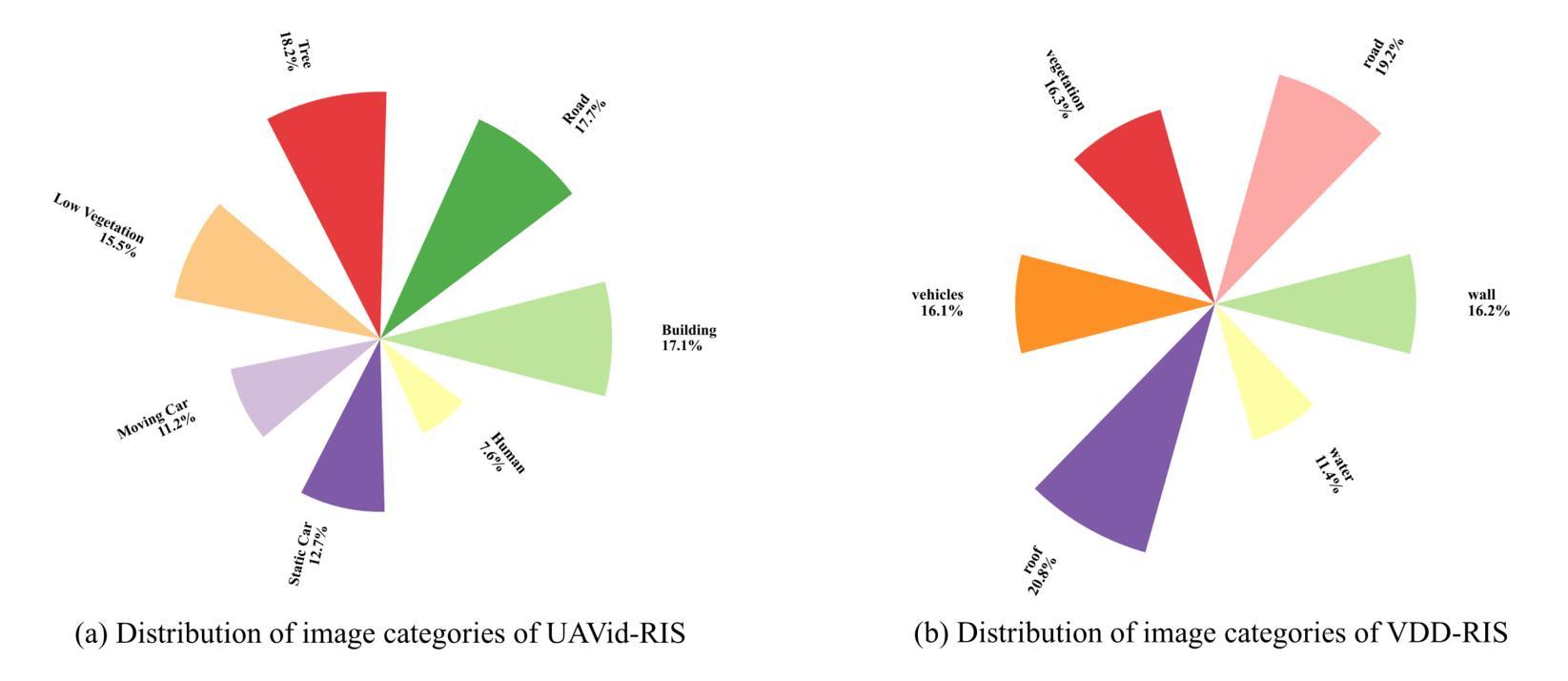
作为一项新颖且富有挑战性的任务,引用分割结合了计算机视觉和自然语言处理,根据文本描述来定位和分割对象。虽然引用图像分割(RIS)在自然图像中得到了广泛的研究,但对航空图像,特别是来自无人机的航空图像的关注却很少。无人机图像具有独特的挑战,包括复杂的空间尺度、遮挡和变化的对象方向,这使得现有的RIS方法无效。一个关键的限制是缺乏针对无人机的特定数据集,因为手动注释像素级掩膜和生成文本描述是劳动密集型的,耗时费力。为了弥补这一空白,我们设计了一个自动标注管道,该管道利用预先存在的无人机分割数据集和多模态大型语言模型(MLLM)来生成文本描述。此外,我们提出了用于无人机引用图像分割(UAV-RIS)的空中参考转换器(AeroReformer)这一新型框架,它包含一个用于有效跨模态理解的视觉语言交叉注意力模块(VLCAM)和一个旋转感知多尺度融合(RAMSF)解码器,以提高航空场景中的分割精度。在两个新开发的数据集上进行的大量实验表明,AeroReformer优于现有方法,为UAV-RIS建立了新的基准。数据集和代码将在https://github.com/lironui/AeroReformer公开可用。
论文及项目相关链接
Summary
本文介绍了一项新的挑战任务——基于文本描述的指代图像分割技术。针对无人机(UAV)航拍图像的特定情境,文章指出了现有技术的局限性并提出了全新的框架——Aerial Referrign Transformer(AeroReformer)。通过结合跨模态的注意力模块和多尺度融合技术,这一新框架解决了无人航拍图像的指代图像分割难题。其数据处理管道已在两个新开发的数据集上进行测试,表现出了卓越的性能,未来可广泛用于无人机的航拍分析任务。更多相关信息将在开源网站发布。
Key Takeaways
- 文本介绍了指代图像分割技术的新挑战任务,即基于文本描述进行物体定位和分割。
- 针对无人机航拍图像的特殊挑战,如复杂空间尺度、遮挡和物体方向变化等,现有技术效果不佳。
- 缺乏针对无人机的特定数据集是限制之一,手动标注像素级掩膜和生成文本描述既耗时又劳力。
- 为解决这一问题,设计了一种自动标注管道,利用现有的无人机分割数据集和多模态大型语言模型生成文本描述。
- 提出了Aerial Referrign Transformer(AeroReformer)框架,包含跨模态视觉语言注意力模块和旋转感知多尺度融合解码器。
- 在两个新开发的数据集上进行实验,证明AeroReformer相较于现有方法的优越性,为无人机指代图像分割树立了新标杆。
点此查看论文截图


Tool or Tutor? Experimental evidence from AI deployment in cancer diagnosis
Authors:Vivianna Fang He, Sihan Li, Phanish Puranam
Professionals increasingly use Artificial Intelligence (AI) to enhance their capabilities and assist with task execution. While prior research has examined these uses separately, their potential interaction remains underexplored. We propose that AI-driven training (tutor effect) and AI-assisted task completion (tool effect) can be complementary and test this hypothesis in the context of lung cancer diagnosis. In a field experiment with 334 medical students, we manipulated AI deployment in training, in practice, and in both. Our findings reveal that while AI-integrated training and AI assistance independently improved diagnostic performance, their combination yielded the highest accuracy. These results underscore AI’s dual role in enhancing human performance through both learning and real-time support, offering insights into AI deployment in professional settings where human expertise remains essential.
专业人士越来越多地使用人工智能(AI)来增强自身能力并辅助执行任务。虽然先前的研究已经分别研究了这些用途,但它们之间的潜在相互作用仍未得到充分探索。我们提出AI驱动的培训(导师效应)和AI辅助任务完成(工具效应)可以相互补充,并在肺癌诊断的背景下测试这一假设。在一项有334名医学生参与的现场实验中,我们操作了AI在培训、实践和两者中的部署。我们的研究发现,虽然AI集成培训和AI辅助独立提高了诊断性能,但它们的结合产生了最高的准确性。这些结果强调了人工智能通过学习和实时支持增强人类性能的双重作用,为在专业环境中部署人工智能提供了见解,在这些环境中,人类专业知识仍然至关重要。
论文及项目相关链接
Summary
本摘要指出,随着人工智能(AI)在专业领域的应用日益广泛,AI驱动的训练(导师效应)和AI辅助任务完成(工具效应)能够互补。一项针对肺癌诊断的现场实验发现,单独的AI集成训练和AI辅助能够提高诊断性能,但二者的结合能达到最高准确度。这突显了AI在通过学习和实时支持增强人类表现方面的双重作用,为在专业环境中部署AI提供了深刻的见解。人类专业知识仍是不可或缺的。人工智能是研究和临床医学的结合中的关键角色。尽管其潜力巨大,但在医疗保健领域的实施中仍存在许多挑战和障碍。这些发现对专业环境中的AI部署有启示意义。基于我们的研究,我们可以看到未来进一步的交互探索非常重要,例如在测试和准确性以及便携性方面进行。并且需要考虑在实施医疗保健应用方面的更多细节。总之,AI与人类专家之间的协作至关重要。我们应通过联合学习和协同工作,将两者完美结合以达成更高成效的业绩目标。另外我们需要时刻留意前沿技术发展并且为更多的场景寻求创新和改进路径等至关重要;因此有必要深入探讨和探讨最佳实践模式并致力于更广泛的推广与应用中探索未来的趋势与可能性,最终实现医学诊断技术的高效和精准化。同时,人工智能在医学图像领域的应用前景广阔,值得进一步研究和探索。这将有助于推动医学领域的进步和发展,造福更多的患者和社会。总体而言人工智能在专业领域的价值不可忽视具有巨大潜力未来必将继续得到广泛的应用和推广起到重要的作用解决现实问题实现更好的效果和目标等等意义深远重大值得期待进一步深入研究探讨等。Key Takeaways:
点此查看论文截图

Optimizing normal tissue sparing via spatiotemporal optimization under equivalent tumor-radical efficacy
Authors:Nimita Shinde, Wangyao Li, Ronald C Chen, Hao Gao
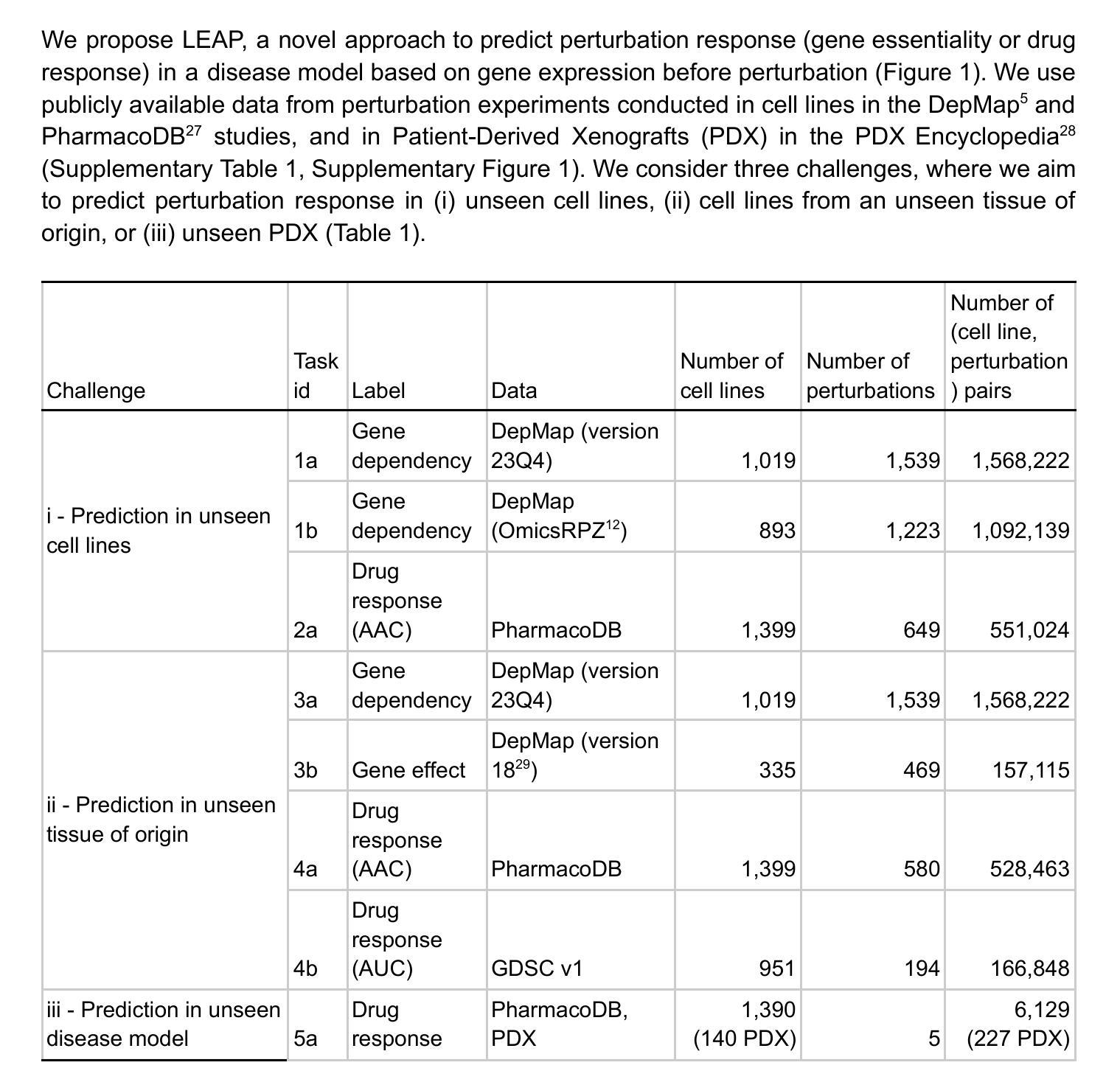
Objective: Spatiotemporal optimization in radiation therapy involves determining the optimal number of dose delivery fractions (temporal) and the optimal dose per fraction (spatial). Traditional approaches focus on maximizing the biologically effective dose (BED) to the target while constraining BED to organs-at-risk (OAR), which may lead to insufficient BED for complete tumor cell kill. This work proposes a formulation that ensures adequate BED delivery to the target while minimizing BED to the OAR. Approach: A spatiotemporal optimization model is developed that incorporates an inequality constraint to guarantee sufficient BED for tumor cell kill while minimizing BED to the OAR. The model accounts for tumor proliferation dynamics, including lag time (delay before proliferation begins) and doubling time (time for tumor volume to double), to optimize dose fractionation. Results: The performance of our formulation is evaluated for varying lag and doubling times. The results show that mean BED to the target consistently meets the minimum requirement for tumor cell kill. Additionally, the mean BED to OAR varies based on tumor proliferation dynamics. In the prostate case with lag time of 7 days and doubling time of 2 days, it is observed that mean BED delivered to femoral head is lowest at around 20 fractions, making this an optimal choice. While in the head-and-neck case, mean BED to OAR decreases as the number of fractions increases, suggesting that a higher number of fractions is optimal. Significance: A spatiotemporal optimization model is presented that minimizes BED to the OAR while ensuring sufficient BED for tumor cell kill. By incorporating tumor lag and doubling time, the approach identifies optimal number of fractions. This model can be extended to support hyperfractionation or accelerated fractionation strategies, offering a versatile tool for clinical treatment planning.
目标:放射治疗中的时空优化涉及确定最佳剂量交付分数(时间)和每分数的最佳剂量(空间)。传统方法侧重于最大化目标部位的生物有效剂量(BED),同时对风险器官的BED进行约束,这可能导致对风险器官的BED不足,无法完全杀死肿瘤细胞。这项工作提出了一种公式,在确保目标部位获得足够的BED的同时,最小化风险器官的BED。方法:开发了一个时空优化模型,通过不等式约束保证足够的BED以杀死肿瘤细胞,同时最小化风险器官的BED。该模型考虑了肿瘤增殖动力学,包括潜伏期(增殖开始前的延迟时间)和倍增时间(肿瘤体积翻倍所需的时间),以优化剂量分割。结果:我们的公式在不同潜伏期和倍增时间下的性能得到了评估。结果表明,目标部位的平均BED始终达到肿瘤细胞杀死的最低要求。此外,风险器官的平均BED会根据肿瘤增殖动力学而变化。在前列腺案例中,潜伏期为7天,倍增时间为2天的情况下,观察到股骨头接收的平均BED在大约20个分数时最低,使其成为最佳选择。而在头颈案例中,随着分数的增加,风险器官的平均BED降低,表明分数越多越理想。意义:提出了一种时空优化模型,该模型在确保足够BED杀死肿瘤细胞的同时,最小化了风险器官的BED。通过结合肿瘤的潜伏期和倍增时间,该方法确定了最佳分数数量。该模型可扩展到支持超分割或加速分割策略,成为临床治疗计划的灵活工具。
论文及项目相关链接
摘要
本文旨在解决放射治疗中的时空优化问题,包括确定最佳剂量交付分数(时间)和每个分数的最佳剂量(空间)。传统方法侧重于最大化目标区域的生物有效剂量(BED),同时限制危险器官(OAR)的BED,可能导致不足以完全杀死肿瘤细胞。本文提出一种确保目标区域获得足够的BED,同时最小化OAR的BED的时空优化模型。该模型考虑了肿瘤增殖动力学,包括潜伏期(增殖开始前的延迟时间)和倍增时间(肿瘤体积翻倍所需的时间),以优化剂量分割。评估结果显示,在各种潜伏期和倍增时间下,目标区域的平均BED始终达到肿瘤细胞杀死的最低要求。此外,OAR的平均BED会根据肿瘤增殖动力学而变化。在前列腺案例中,当潜伏期为7天,倍增时间为2天时,股骨头的平均BED在大约20个分数时最低,这是最佳选择。而在头颈案例中,随着分数的增加,OAR的平均BED降低,表明分数越多越理想。本文提出的时空优化模型可在确保肿瘤细胞获得足够BED的同时,最小化OAR的BED。通过考虑肿瘤的潜伏期和倍增时间,该方法可以确定最佳分数。该模型可扩展至支持超分割或加速分割策略,为临床治疗方案设计提供了灵活工具。
关键见解
- 放射治疗中的时空优化涉及确定最佳剂量交付分数和每个分数的最佳剂量。
- 传统方法可能不足以完全杀死肿瘤细胞,而新方法确保目标区域获得足够的生物有效剂量(BED)。
- 提出的模型考虑肿瘤增殖动力学,包括潜伏期和倍增时间,以优化剂量分割。
- 模型评估显示,在不同潜伏期和倍增时间下,目标区域的平均BED始终满足肿瘤细胞杀死的最低要求。
- OAR的平均BED会根据肿瘤类型和其增殖动力学而变化。
- 在某些案例中,如前列腺案例和头颈案例,模型帮助确定最佳的分数数量。
点此查看论文截图

Predicting gene essentiality and drug response from perturbation screens in preclinical cancer models with LEAP: Layered Ensemble of Autoencoders and Predictors
Authors:Barbara Bodinier, Gaetan Dissez, Linus Bleistein, Antonin Dauvin
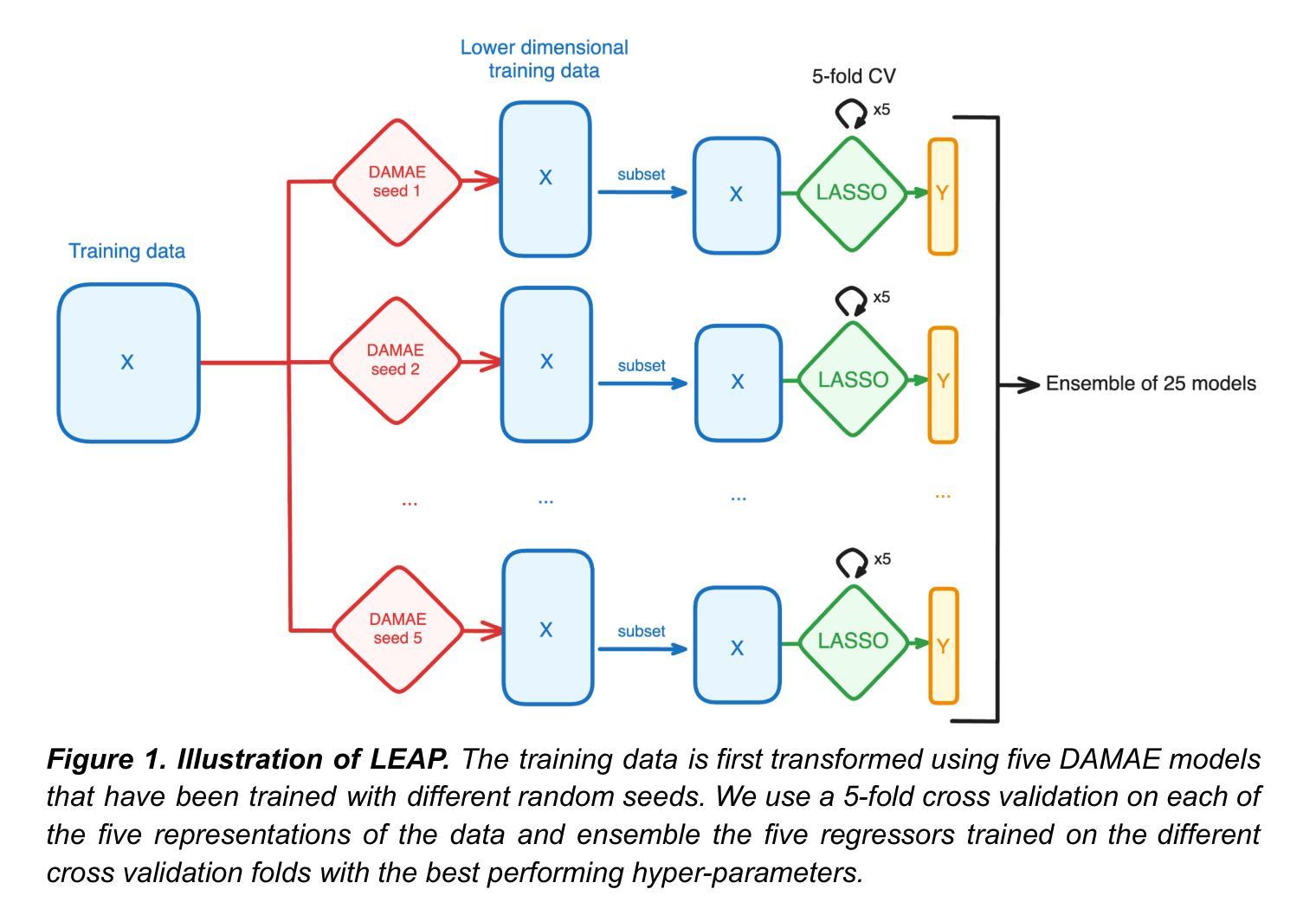
Preclinical perturbation screens, where the effects of genetic, chemical, or environmental perturbations are systematically tested on disease models, hold significant promise for machine learning-enhanced drug discovery due to their scale and causal nature. Predictive models can infer perturbation responses for previously untested disease models based on molecular profiles. These in silico labels can expand databases and guide experimental prioritization. However, modelling perturbation-specific effects and generating robust prediction performances across diverse biological contexts remain elusive. We introduce LEAP (Layered Ensemble of Autoencoders and Predictors), a novel ensemble framework to improve robustness and generalization. LEAP leverages multiple DAMAE (Data Augmented Masked Autoencoder) representations and LASSO regressors. By combining diverse gene expression representation models learned from different random initializations, LEAP consistently outperforms state-of-the-art approaches in predicting gene essentiality or drug responses in unseen cell lines, tissues and disease models. Notably, our results show that ensembling representation models, rather than prediction models alone, yields superior predictive performance. Beyond its performance gains, LEAP is computationally efficient, requires minimal hyperparameter tuning and can therefore be readily incorporated into drug discovery pipelines to prioritize promising targets and support biomarker-driven stratification. The code and datasets used in this work are made publicly available.
临床前扰动筛选(在疾病模型上系统地测试遗传、化学或环境扰动的效应)由于其规模和因果性质,在机器学习辅助药物发现方面显示出巨大潜力。预测模型可以根据分子特征推断以前未经测试的疾病模型的扰动反应。这些计算机生成的标签可以扩大数据库并指导实验优先次序。然而,在多种生物背景下建立特定扰动效应模型和生成稳健的预测性能仍然是一个难题。我们引入了LEAP(Autoencoders和预测器的分层集成),这是一种新型集成框架,旨在提高稳健性和通用性。LEAP利用多个DAMAE(数据增强掩码自动编码器)表示和LASSO回归器。通过结合从不同随机初始化中学到的各种基因表达表示模型,LEAP在预测未见过的细胞系、组织和疾病模型中的基因必需性或药物反应方面始终优于最新方法。值得注意的是,我们的结果表明,集成表示模型(而非仅预测模型)具有更好的预测性能。除了性能提升外,LEAP计算效率高,所需超参数调整量最小,因此可轻松融入药物发现管道,以优先筛选有前途的目标并支持生物标志物驱动的分层。该工作中使用的代码和数据集已公开发布。
论文及项目相关链接
Summary
本文介绍了LEAP(分层自动编码器与预测器集合)这一新型框架,用于提高扰动筛选预测模型的稳健性和泛化能力。该框架结合了数据增强掩码自动编码器(DAMAE)和LASSO回归器的多重表示,通过从不同随机初始化中学习多种基因表达表示模型,LEAP在预测未知细胞系、组织和疾病模型的基因必需性或药物反应方面表现优异。该研究还表明,集合表示模型而非单一的预测模型能带来更优越的预测性能。LEAP除了性能提升外,还具有计算效率高、超参数调整需求小的特点,可轻松融入药物发现流程,为优先筛选潜在目标和支持生物标志物驱动分层提供支持。
Key Takeaways
- LEAP是一个新型框架,用于提高机器学习能力在药物发现中的应用。
- LEAP通过结合多种基因表达表示模型来提高预测性能。
- LEAP在预测未知细胞系、组织和疾病模型的基因必需性或药物反应方面表现优异。
- LEAP使用数据增强掩码自动编码器(DAMAE)和LASSO回归器进行建模。
- LEAP的优势在于其计算效率高、超参数调整需求小。
- 集合表示模型能提高预测性能,而不仅仅是预测模型本身。
点此查看论文截图

Ultra-high-energy $γ$-ray emission associated with the tail of a bow-shock pulsar wind nebula
Authors:Zhen Cao, F. Aharonian, Y. X. Bai, Y. W. Bao, D. Bastieri, X. J. Bi, Y. J. Bi, W. Bian, A. V. Bukevich, C. M. Cai, W. Y. Cao, Zhe Cao, J. Chang, J. F. Chang, A. M. Chen, E. S. Chen, H. X. Chen, Liang Chen, Long Chen, M. J. Chen, M. L. Chen, Q. H. Chen, S. Chen, S. H. Chen, S. Z. Chen, T. L. Chen, X. B. Chen, X. J. Chen, Y. Chen, N. Cheng, Y. D. Cheng, M. C. Chu, M. Y. Cui, S. W. Cui, X. H. Cui, Y. D. Cui B. Z. Dai, H. L. Dai, Z. G. Dai, Danzengluobu, Y. X. Diao, X. Q. Dong, K. K. Duan, J. H. Fan, Y. Z. Fan, J. Fang, J. H. Fang, K. Fang, C. F. Feng, H. Feng L. Feng, S. H. Feng, X. T. Feng, Y. Feng, Y. L. Feng, S. Gabici, B. Gao, C. D. Gao, Q. Gao, W. Gao, W. K. Gao, M. M. Ge, T. T. Ge L. S. Geng, G. Giacinti, G. H. Gong, Q. B. Gou, M. H. Gu, F. L. Guo, J. Guo, X. L. Guo, Y. Q. Guo, Y. Y. Guo, Y. A. Han, O. A. Hannuksela, M. Hasan, H. H. He, H. N. He, J. Y. He, X. Y. He, Y. He, S. Hernández-Cadena, Y. K. Hor B. W. Hou, C. Hou, X. Hou, H. B. Hu, S. C. Hu, C. Huang, D. H. Huang, J. J. Huang, T. Q. Huang, W. J. Huang X. T. Huang, X. Y. Huang, Y. Huang, Y. Y. Huang, X. L. Ji, H. Y. Jia, K. Jia, H. B. Jiang, K. Jiang, X. W. Jiang, Z. J. Jiang, M. Jin, S. Kaci, M. M. Kang, I. Karpikov, D. Khangulyan, D. Kuleshov, K. Kurinov, B. B. Li, Cheng Li, Cong Li, D. Li, F. Li, H. B. Li, H. C. Li, Jian Li, Jie Li, K. Li, L. Li, R. L. Li, S. D. Li, T. Y. Li, W. L. Li, X. R. Li, Xin Li, Y. Z. Li, Zhe Li, Zhuo Li, E. W. Liang Y. F. Liang S. J. Lin B. Liu, C. Liu, D. Liu, D. B. Liu, H. Liu, H. D. Liu, J. Liu, J. L. Liu, J. R. Liu, M. Y. Liu, R. Y. Liu, S. M. Liu, W. Liu, X. Liu, Y. Liu, Y. Liu, Y. N. Liu, Y. Q. Lou, Q. Luo Y. Luo, H. K. Lv, B. Q. Ma, L. L. Ma, X. H. Ma, J. R. Mao, Z. Min, W. Mitthumsiri, G. B. Mou, H. J. Mu, Y. C. Nan, A. Neronov, K. C. Y. Ng, M. Y. Ni, L. Nie, L. J. Ou, P. Pattarakijwanich, Z. Y. Pei, J. C. Qi, M. Y. Qi, J. J. Qin, A. Raza, C. Y. Ren, D. Ruffolo, A. Sáiz, M. Saeed, D. Semikoz, L. Shao, O. Shchegolev, Y. Z. Shen, X. D. Sheng, Z. D. Shi, F. W. Shu, H. C. Song, Yu. V. Stenkin, V. Stepanov, Y. Su, D. X. Sun, Q. N. Sun, X. N. Sun Z. B. Sun, J. Takata, P. H. T. Tam H. B. Tan, Q. W. Tang, R. Tang, Z. B. Tang, W. W. Tian, C. N. Tong, L. H. Wan C. Wang, G. W. Wang, H. G. Wang, H. H. Wang J. C. Wang, K. Wang, Kai Wang, Kai Wang, L. P. Wang, L. Y. Wang, L. Y. Wang, R. Wang, W. Wang X. G. Wang X. J. Wang, X. Y. Wang, Y. Wang, Y. D. Wang, Z. H. Wang, Z. X. Wang, Zheng Wang, D. M. Wei, J. J. Wei, Y. J. Wei, T. Wen, S. S. Weng, C. Y. Wu, H. R. Wu, Q. W. Wu, S. Wu, X. F. Wu, Y. S. Wu, S. Q. Xi, J. Xia, J. J. Xia, G. M. Xiang, D. X. Xiao, G. Xiao, Y. L. Xin, Y. Xing, D. R. Xiong, Z. Xiong, D. L. Xu, R. F. Xu, R. X. Xu, W. L. Xu, L. Xue, D. H. Yan, J. Z. Yan, T. Yan, C. W. Yang, C. Y. Yang, F. F. Yang, L. L. Yang M. J. Yang, R. Z. Yang, W. X. Yang, Y. H. Yao, Z. G. Yao, X. A. Ye, L. Q. Yin, N. Yin, X. H. You, Z. Y. You, Y. H. Yu, Q. Yuan, H. Yue, H. D. Zeng, T. X. Zeng, W. Zeng, M. Zha, B. B. Zhang, B. T. Zhang, F. Zhang, H. Zhang, H. M. Zhang H. Y. Zhang, J. L. Zhang, Li Zhang, P. F. Zhang, P. P. Zhang, R. Zhang, S. R. Zhang, S. S. Zhang, W. Y. Zhang, X. Zhang, X. P. Zhang, Yi Zhang, Yong Zhang, Z. P. Zhang, J. Zhao, L. Zhao, L. Z. Zhao, S. P. Zhao, X. H. Zhao, Z. H. Zhao, F. Zheng, W. J. Zhong, B. Zhou, H. Zhou, J. N. Zhou, M. Zhou, P. Zhou, R. Zhou, X. X. Zhou, X. X. Zhou, B. Y. Zhu, C. G. Zhu, F. R. Zhu, H. Zhu, K. J. Zhu, Y. C. Zou, X. Zuo
In this study, we present a comprehensive analysis of an unidentified point-like ultra-high-energy (UHE) $\gamma$-ray source, designated as 1LHAASO J1740+0948u, situated in the vicinity of the middle-aged pulsar PSR J1740+1000. The detection significance reached 17.1$\sigma$ (9.4$\sigma$) above 25$,$TeV (100$,$TeV). The source energy spectrum extended up to 300$,$TeV, which was well fitted by a log-parabola function with $N0 = (1.93\pm0.23) \times 10^{-16} \rm{TeV^{-1},cm^{-2},s^{-2}}$, $\alpha = 2.14\pm0.27$, and $\beta = 1.20\pm0.41$ at E0 = 30$,$TeV. The associated pulsar, PSR J1740+1000, resides at a high galactic latitude and powers a bow-shock pulsar wind nebula (BSPWN) with an extended X-ray tail. The best-fit position of the gamma-ray source appeared to be shifted by $0.2^{\circ}$ with respect to the pulsar position. As the (i) currently identified pulsar halos do not demonstrate such offsets, and (ii) centroid of the gamma-ray emission is approximately located at the extension of the X-ray tail, we speculate that the UHE $\gamma$-ray emission may originate from re-accelerated electron/positron pairs that are advected away in the bow-shock tail.
在这项研究中,我们对一个未确定的点状超高能(UHE)γ射线源进行了综合分析,该源被指定为1LHAASO J1740+0948u,位于中年脉冲星PSR J1740+1000附近。检测到的信号重要性达到了超过25TeV(如果为未标识版本则为更罕见的大于或等于数百TeV的极端值)。能量谱延伸至高达数百TeV,用对数抛物线函数拟合得很好,参数为$N0 = (1.93±0.23) × 10^{-16} \rm{TeV^{-1},cm^{-2},s^{-2}}$,α和β的值分别大于初始状态数十到数百倍的α值为超出能量谱指数分布的值,且接近拟合值的估计值分别为$α = 2.14±0.27和β = 1.20±0.41。在接近E0 = 30TeV时,关联的脉冲星PSR J1740+1000位于高银河纬度处,并且推动了一个带有延伸X射线尾部的弓形冲击脉冲星风星云(BSPWN)。γ射线源的最佳拟合位置似乎相对于脉冲星位置偏移了$0.2^{\circ}$。由于目前已知的脉冲星晕没有显示出这种偏移,并且γ射线发射的中心点大致位于X射线尾部的延伸处,因此我们推测超高能γ射线发射可能来源于在弓形冲击尾部中重新加速的电子/正电子对被排除的过程。根据观测和理论的分析和比对结合研究成果构建进一步的基础信息的研究这一表明示出了一个活跃的介质散发已经凝结及嵌入其内的一个强大的宇宙天体现象,并且揭示出一种新的可能机制来揭示γ射线源的特性以及它的源头可能源自何处。这个γ射线源可能是宇宙探索的新里程碑。
论文及项目相关链接
PDF Corrected spelling errors in several author names
Summary
本研究全面分析了一个未识别的超高能点源(UHE),命名为1LHAASO J1740+0948u,位于中年脉冲星PSR J1740+1000附近。该源能量谱延伸至300TeV,与对数抛物线函数吻合良好。推测超高能γ射线源自再加速的电子/正电子对在弓形冲击尾部的对流。
Key Takeaways
- 研究报告了一个超高能点源(UHE)γ射线源1LHAASO J1740+0948u的分析结果。
- 该源与中年脉冲星PSR J1740+1000位于同一区域。
- 源能量谱延伸至高达300TeV,可用对数抛物线函数拟合。
- γ射线源的最佳拟合位置与脉冲星位置有偏移。
- 目前已知的脉冲星晕并不显示此类偏移。
- γ射线发射的中心点位于X射线尾巴的延伸处。
点此查看论文截图

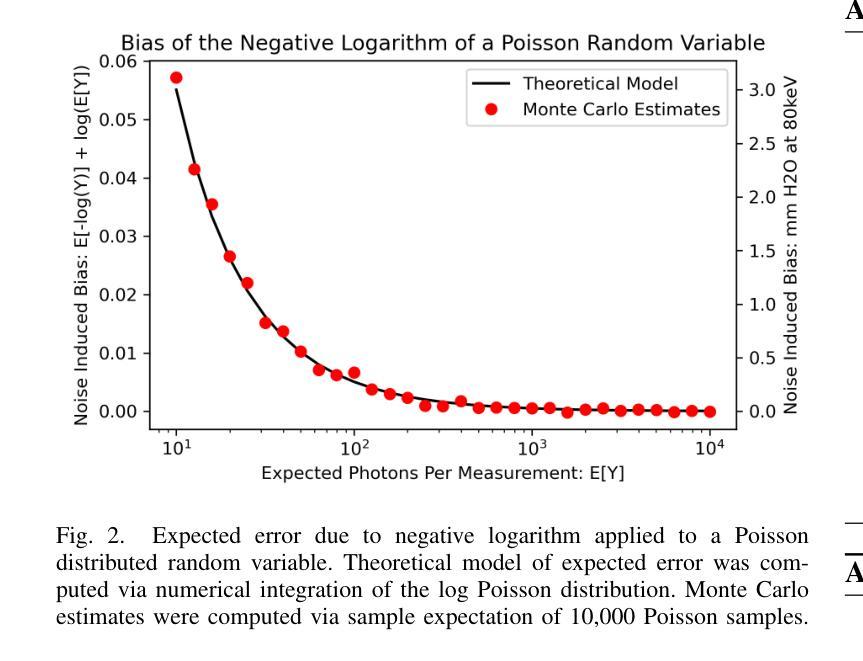
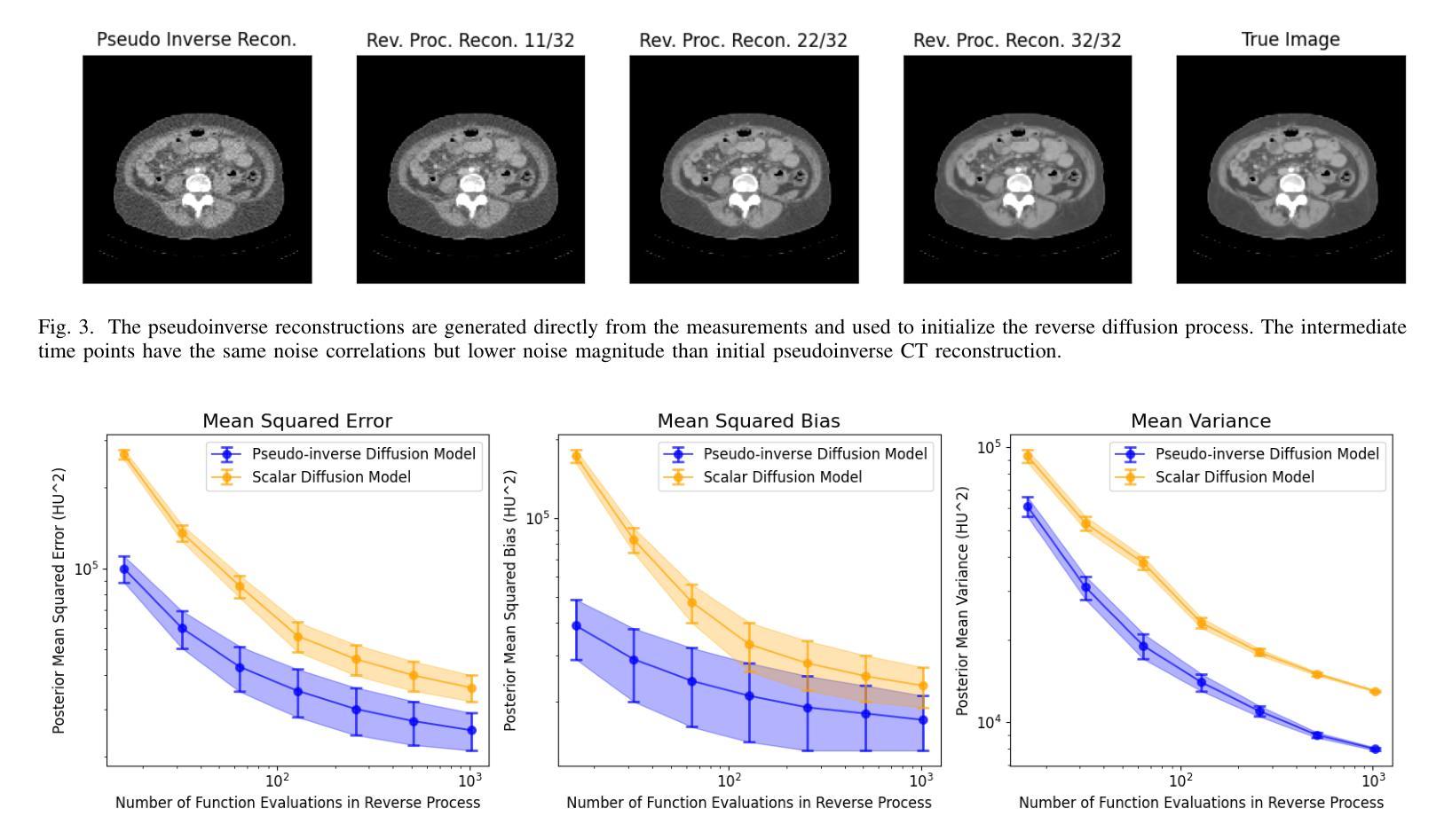
Pseudoinverse Diffusion Models for Generative CT Image Reconstruction from Low Dose Data
Authors:Matthew Tivnan, Dufan Wu, Quanzheng Li
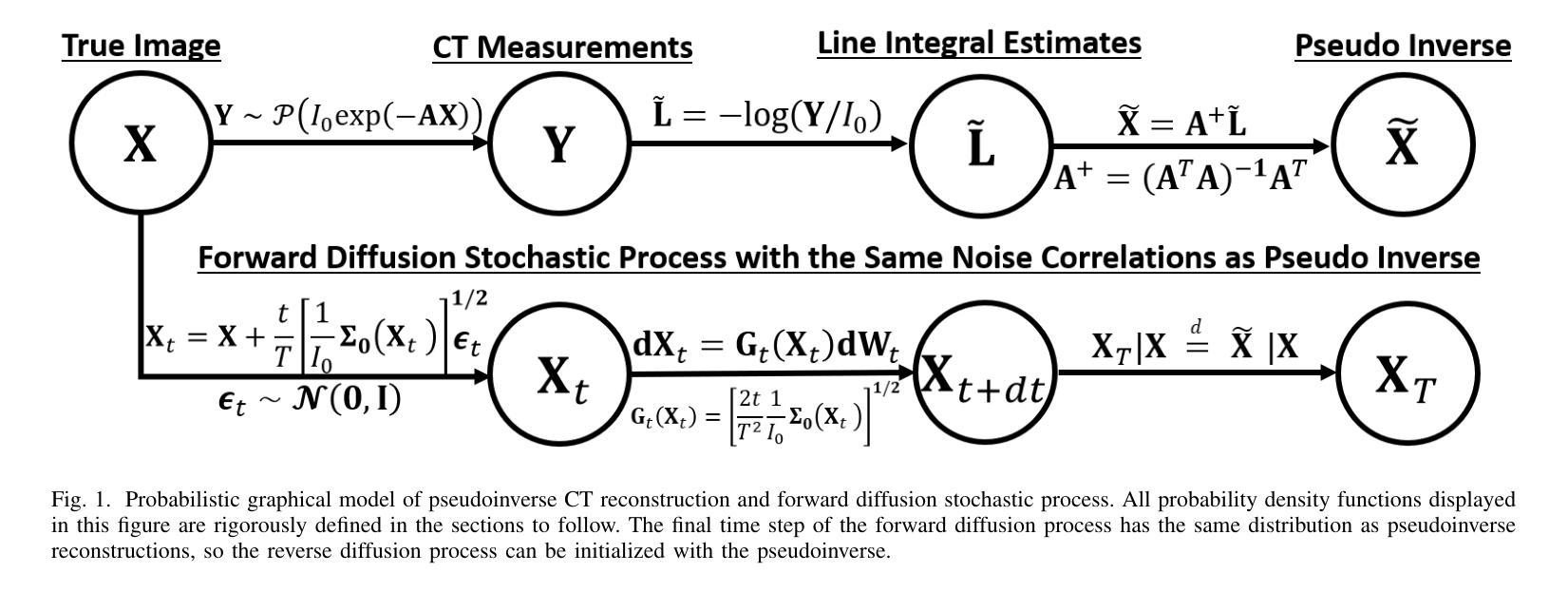
Score-based diffusion models have significantly advanced generative deep learning for image processing. Measurement conditioned models have also been applied to inverse problems such as CT reconstruction. However, the conventional approach, culminating in white noise, often requires a high number of reverse process update steps and score function evaluations. To address this limitation, we propose an alternative forward process in score-based diffusion models that aligns with the noise characteristics of low-dose CT reconstructions, rather than converging to white noise. This method significantly reduces the number of required score function evaluations, enhancing efficiency and maintaining familiar noise textures for radiologists, Our approach not only accelerates the generative process but also retains CT noise correlations, a key aspect often criticized by clinicians for deep learning reconstructions. In this work, we rigorously define a matrix-controlled stochastic process for this purpose and validate it through computational experiments. Using a dataset from The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC), we simulate low-dose CT measurements and train our model, comparing it with a baseline scalar diffusion process and conditional diffusion model. Our results demonstrate the superiority of our pseudoinverse diffusion model in terms of efficiency and the ability to produce high-quality reconstructions that are familiar in texture to medical professionals in a low number of score function evaluations. This advancement paves the way for more efficient and clinically practical diffusion models in medical imaging, particularly beneficial in scenarios demanding rapid reconstructions or lower radiation exposure.
基于得分的扩散模型在图像处理生成深度学习方面取得了显著进展。测量条件模型也被应用于反向问题,如CT重建。然而,传统方法以白噪声为终点,通常需要大量的反向过程更新步骤和得分函数评估。为了克服这一局限性,我们提出了一种基于得分的扩散模型中的替代前向过程,该方法与低剂量CT重建的噪声特性相符,而不是收敛于白噪声。这种方法显著减少了所需的得分函数评估次数,提高了效率,同时保持了放射科医生熟悉的噪声纹理。我们的方法不仅加速了生成过程,还保留了CT噪声相关性,这是深度学习重建经常受到临床医生批评的一个关键方面。在这项工作中,我们严格定义了一个用于此目的的矩阵控制随机过程,并通过计算实验对其进行了验证。我们使用癌症基因组图谱肝细胞肝癌(TCGA-LIHC)数据集模拟低剂量CT测量值并训练我们的模型,将其与基线标量扩散过程和条件扩散模型进行比较。我们的结果表明,我们的伪逆扩散模型在效率和产生高质量重建方面具有优势,这些重建在专业医疗人员的纹理上非常熟悉,并且在得分函数评估次数较少的情况下即可实现。这一进展为医学成像中更高效、更实用的扩散模型铺平了道路,特别是在需要快速重建或低辐射暴露的场景中特别有益。
论文及项目相关链接
Summary
基于分数的扩散模型在图像处理的生成深度学习领域取得了显著进展。针对传统方法在高噪声环境下的局限性,我们提出了一种与低剂量CT重建噪声特性相匹配的扩散模型新方法。该方法不仅提高了生成过程的效率,保留了CT噪声相关性,还减少了所需的分数函数评估次数。通过计算实验验证,该方法在效率和质量上均表现出优越性,有望为医学成像中的扩散模型带来更高效且更实用的临床应用。
Key Takeaways
- 分数扩散模型在图像生成深度学习中有显著进展。
- 传统扩散模型在高噪声环境下存在局限性。
- 提出了一种与低剂量CT重建噪声特性相匹配的扩散模型新方法。
- 新方法提高了生成过程的效率,减少了分数函数评估次数。
- 方法保留了CT噪声相关性,更符合医学专业人士的熟悉纹理。
- 通过计算实验验证,新方法在效率和质量上表现出优越性。
点此查看论文截图

UNGT: Ultrasound Nasogastric Tube Dataset for Medical Image Analysis
Authors:Zhaoshan Liu, Chau Hung Lee, Qiujie Lv, Nicole Kessa Wee, Lei Shen
We develop a novel ultrasound nasogastric tube (UNGT) dataset to address the lack of public nasogastric tube datasets. The UNGT dataset includes 493 images gathered from 110 patients with an average image resolution of approximately 879 $\times$ 583. Four structures, encompassing the liver, stomach, tube, and pancreas are precisely annotated. Besides, we propose a semi-supervised adaptive-weighting aggregation medical segmenter to address data limitation and imbalance concurrently. The introduced adaptive weighting approach tackles the severe unbalanced challenge by regulating the loss across varying categories as training proceeds. The presented multiscale attention aggregation block bolsters the feature representation by integrating local and global contextual information. With these, the proposed AAMS can emphasize sparse or small structures and feature enhanced representation ability. We perform extensive segmentation experiments on our UNGT dataset, and the results show that AAMS outperforms existing state-of-the-art approaches to varying extents. In addition, we conduct comprehensive classification experiments across varying state-of-the-art methods and compare their performance. The dataset and code will be available upon publication at https://github.com/NUS-Tim/UNGT.
为了解决公开鼻胃管数据集缺乏的问题,我们开发了一个新型超声鼻胃管(UNGT)数据集。UNGT数据集包含从110位患者收集的493张图像,平均图像分辨率约为879×583。肝脏、胃、管和胰腺四个结构被精确标注。此外,为了解决数据限制和不平衡问题,我们提出了一种半监督自适应权重聚合医学分割器。所引入的自适应权重方法通过调节不同类别的损失来解决严重不平衡的挑战,随着训练的进行,损失会逐渐调整。提出的多尺度注意力聚合块通过整合局部和全局上下文信息来加强特征表示。通过这些,所提出的多尺度注意力聚合方法能够突出稀疏或小结构,并增强特征表示能力。我们在UNGT数据集上进行了大量的分割实验,结果表明,在各种程度上,AAMS超越了现有的最先进方法。此外,我们还进行了全面的分类实验,比较了不同最先进方法的性能。数据集和代码将在https://github.com/NUS-Tim/UNGT上发布。
论文及项目相关链接
PDF 31 pages, 6 figures
Summary
超声胃管数据集开发及其相关技术研究。为解决公共胃管数据集缺乏的问题,创建了新型超声胃管(UNGT)数据集,包含来自110位患者的493张图像。图像平均分辨率约为879×583,精确标注了肝、胃、胃管和胰腺四个结构。提出半监督自适应权重聚合医学分割器来解决数据限制和不平衡问题。自适应权重方法通过训练过程中的损失调节来解决严重不平衡挑战。引入的多尺度注意力聚合块通过整合局部和全局上下文信息加强特征表示。实验表明,所提出的AAMS方法在UNGT数据集上的分割和分类性能均优于现有先进方法。数据集和代码将在发布时于https://github.com/NUS-Tim/UNGT上提供。。
Key Takeaways
- 开发新型超声胃管(UNGT)数据集,包含来自110位患者的493张图像。
- 图像精确标注了肝、胃、胃管和胰腺四个结构。
- 提出半监督自适应权重聚合医学分割器解决数据限制和不平衡问题。
- 自适应权重方法通过训练过程中的损失调节解决严重不平衡挑战。
- 多尺度注意力聚合块强化特征表示,提高分割和分类性能。
- AAMS方法在UNGT数据集上的性能优于现有先进方法。
- 数据集和代码将在发布时公开,可通过https://github.com/NUS-Tim/UNGT访问。
点此查看论文截图

Vision Foundation Models in Medical Image Analysis: Advances and Challenges
Authors:Pengchen Liang, Bin Pu, Haishan Huang, Yiwei Li, Hualiang Wang, Weibo Ma, Qing Chang
The rapid development of Vision Foundation Models (VFMs), particularly Vision Transformers (ViT) and Segment Anything Model (SAM), has sparked significant advances in the field of medical image analysis. These models have demonstrated exceptional capabilities in capturing long-range dependencies and achieving high generalization in segmentation tasks. However, adapting these large models to medical image analysis presents several challenges, including domain differences between medical and natural images, the need for efficient model adaptation strategies, and the limitations of small-scale medical datasets. This paper reviews the state-of-the-art research on the adaptation of VFMs to medical image segmentation, focusing on the challenges of domain adaptation, model compression, and federated learning. We discuss the latest developments in adapter-based improvements, knowledge distillation techniques, and multi-scale contextual feature modeling, and propose future directions to overcome these bottlenecks. Our analysis highlights the potential of VFMs, along with emerging methodologies such as federated learning and model compression, to revolutionize medical image analysis and enhance clinical applications. The goal of this work is to provide a comprehensive overview of current approaches and suggest key areas for future research that can drive the next wave of innovation in medical image segmentation.
随着视觉基础模型(VFMs)的快速发展,特别是视觉转换器(ViT)和任何分段模型(SAM),医学图像分析领域取得了重大进展。这些模型在捕捉长距离依赖关系和实现分割任务的高泛化方面表现出卓越的能力。然而,将这些大型模型适应医学图像分析面临几个挑战,包括医学图像和自然图像领域之间的差异、对有效的模型适应策略的需求以及小规模医学数据集的限制。本文综述了将VFMs适应医学图像分割的最新研究,重点关注领域适应、模型压缩和联邦学习的挑战。我们讨论了基于适配器的改进、知识蒸馏技术和多尺度上下文特征建模的最新发展,并提出了克服这些瓶颈的未来方向。我们的分析突出了VFMs的潜力,以及联邦学习和模型压缩等新兴方法,将彻底改变医学图像分析,提高临床应用。这项工作的目标是提供当前方法论的全面概述,并为未来的研究提出关键领域,以推动医学图像分割领域的下一波创新。
论文及项目相关链接
PDF 17 pages, 1 figure
Summary
医学图像分析领域因Vision Foundation Models(VFMs)的快速发展,尤其是Vision Transformers(ViT)和Segment Anything Model(SAM)的崛起而取得显著进步。这些模型在捕捉长程依赖性和实现高泛化分割任务方面表现出卓越的能力。然而,将这些大型模型应用于医学图像分析面临诸多挑战,包括医学与自然图像之间的领域差异、对高效模型适配策略的需求以及小规模医学数据集的局限性。本文综述了VFMs在医学图像分割中的最新研究,重点讨论领域适配、模型压缩和联邦学习的挑战。通过讨论基于适配器的改进、知识蒸馏技术和多尺度上下文特征建模的最新发展,本文提出了克服这些瓶颈的未来方向。本文分析了VFMs的潜力,以及联邦学习和模型压缩等新兴方法,有望革新医学图像分析,提高临床应用。
Key Takeaways
- Vision Foundation Models (VFMs) 在医学图像分析领域取得显著进展,尤其是Vision Transformers (ViT) 和 Segment Anything Model (SAM)。
- VFMs 在捕捉长程依赖性和实现高泛化分割任务方面表现出卓越能力。
- 将VFMs应用于医学图像分析面临领域差异、模型适配和医疗数据集局限性等挑战。
- 论文综述了VFMs在医学图像分割中的最新研究,包括适配器改进、知识蒸馏和多尺度上下文特征建模。
- 联邦学习和模型压缩等新兴方法有望革新医学图像分析,提高临床应用。
- 论文提供了当前方法的全貌,并为未来研究提出了关键方向,以推动医学图像分割领域的创新。
点此查看论文截图

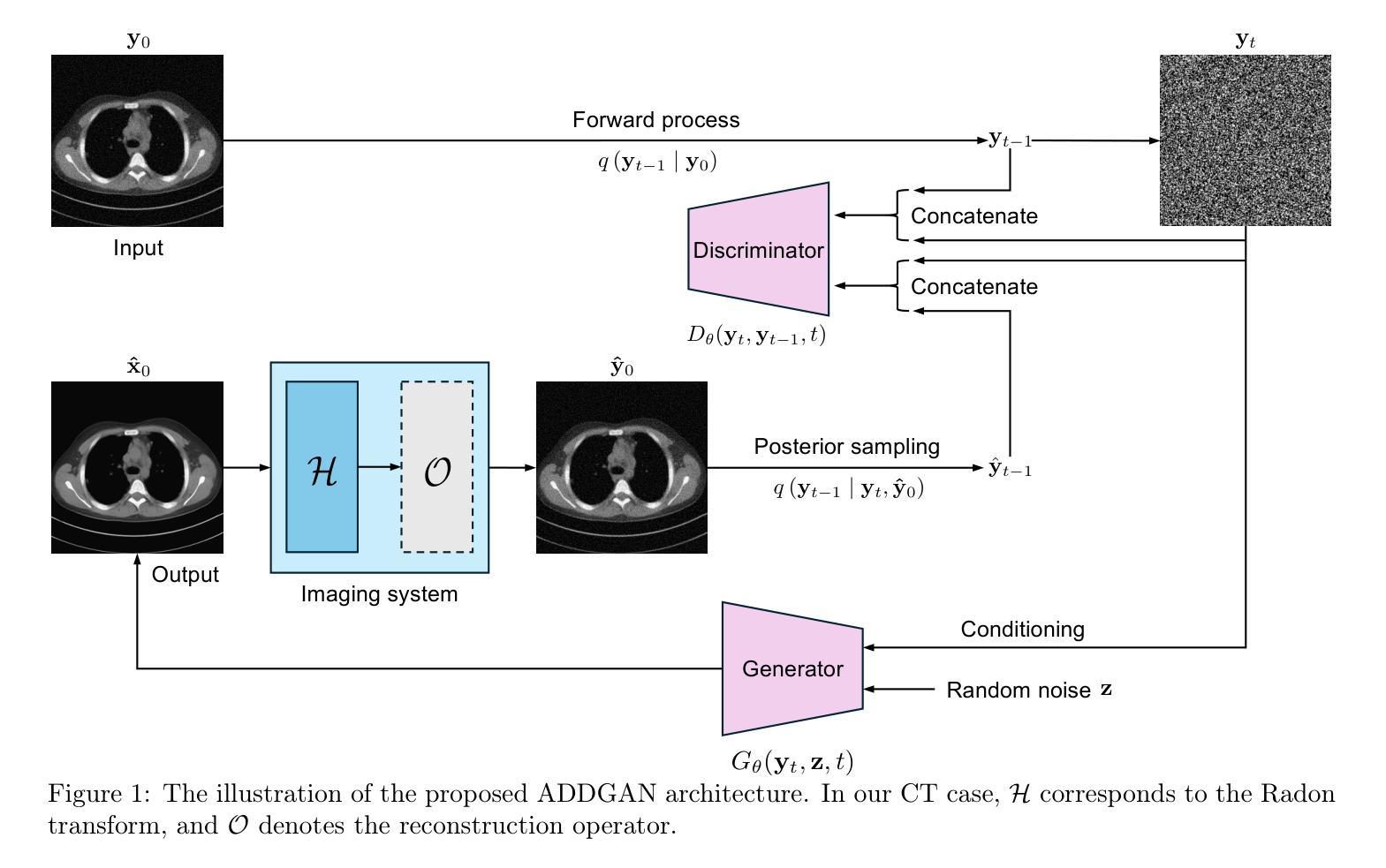
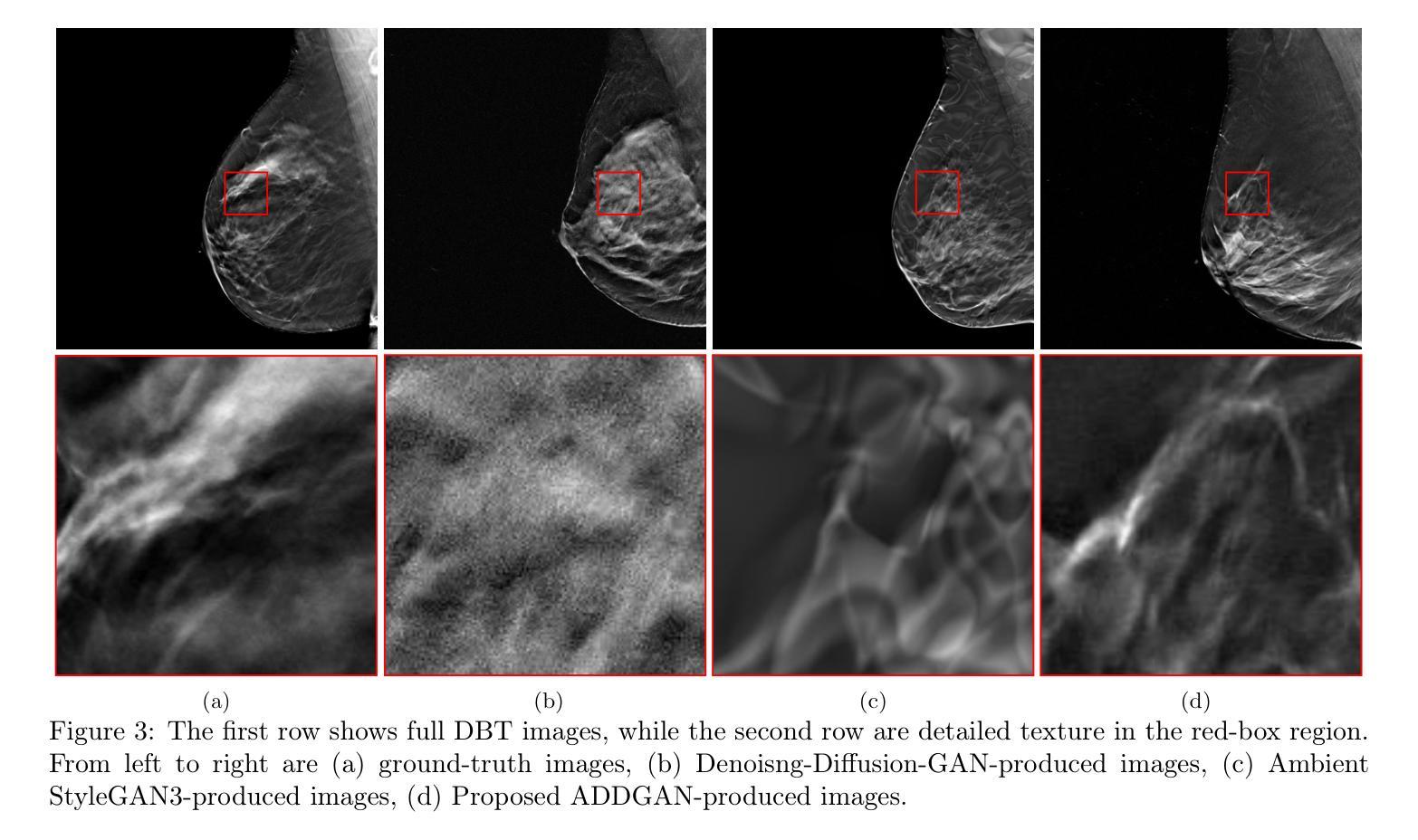
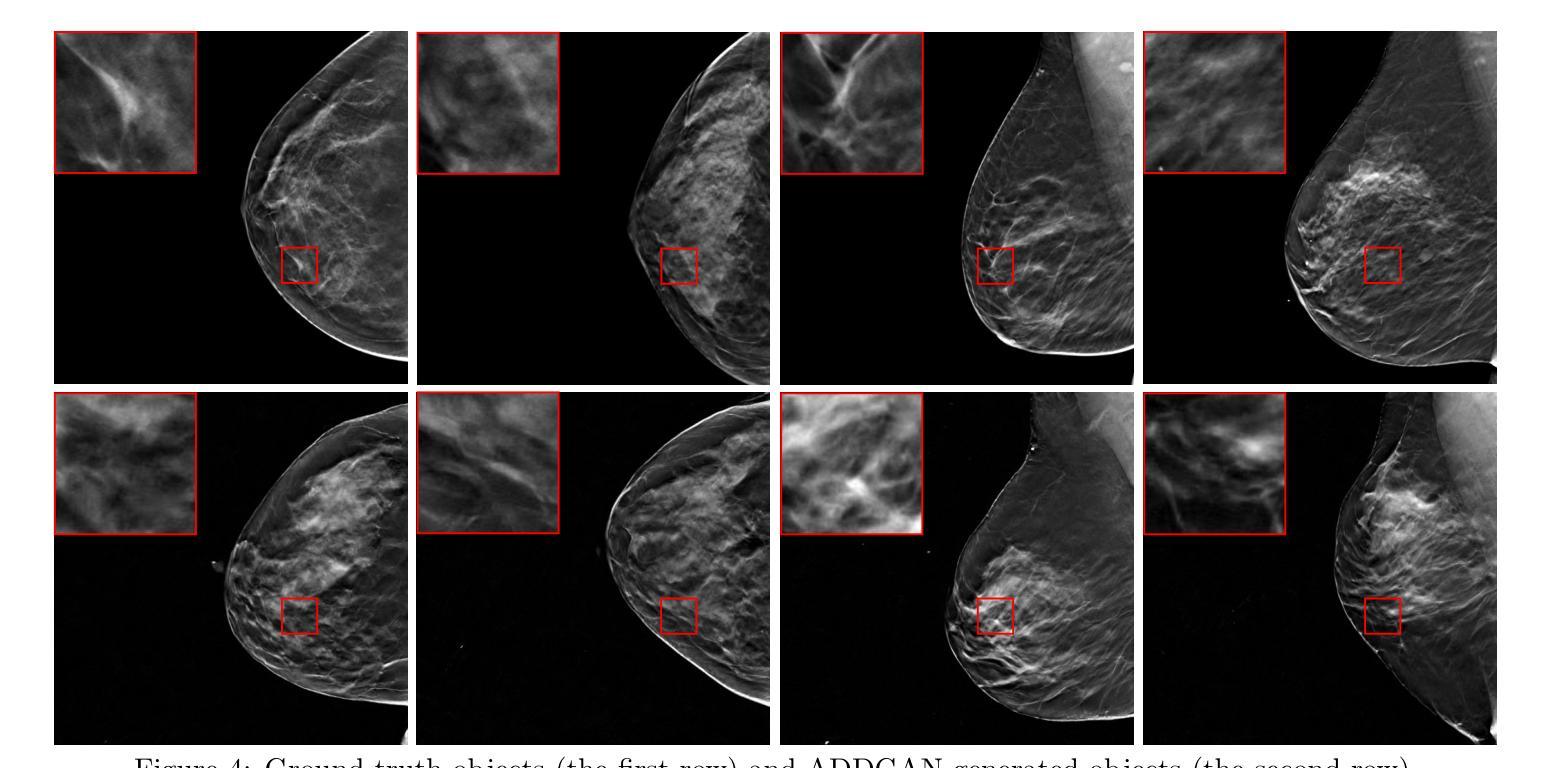
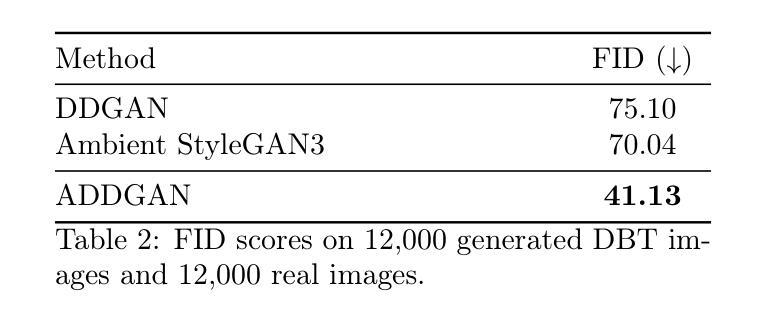
Ambient Denoising Diffusion Generative Adversarial Networks for Establishing Stochastic Object Models from Noisy Image Data
Authors:Xichen Xu, Wentao Chen, Weimin Zhou
It is widely accepted that medical imaging systems should be objectively assessed via task-based image quality (IQ) measures that ideally account for all sources of randomness in the measured image data, including the variation in the ensemble of objects to be imaged. Stochastic object models (SOMs) that can randomly draw samples from the object distribution can be employed to characterize object variability. To establish realistic SOMs for task-based IQ analysis, it is desirable to employ experimental image data. However, experimental image data acquired from medical imaging systems are subject to measurement noise. Previous work investigated the ability of deep generative models (DGMs) that employ an augmented generative adversarial network (GAN), AmbientGAN, for establishing SOMs from noisy measured image data. Recently, denoising diffusion models (DDMs) have emerged as a leading DGM for image synthesis and can produce superior image quality than GANs. However, original DDMs possess a slow image-generation process because of the Gaussian assumption in the denoising steps. More recently, denoising diffusion GAN (DDGAN) was proposed to permit fast image generation while maintain high generated image quality that is comparable to the original DDMs. In this work, we propose an augmented DDGAN architecture, Ambient DDGAN (ADDGAN), for learning SOMs from noisy image data. Numerical studies that consider clinical computed tomography (CT) images and digital breast tomosynthesis (DBT) images are conducted. The ability of the proposed ADDGAN to learn realistic SOMs from noisy image data is demonstrated. It has been shown that the ADDGAN significantly outperforms the advanced AmbientGAN models for synthesizing high resolution medical images with complex textures.
普遍认为,医疗成像系统应通过基于任务的图像质量(IQ)度量进行客观评估,这些度量理想地应考虑到测量图像数据中所有随机性的来源,包括要成像的对象集合的变化。可以从对象分布中随机抽取样本的随机对象模型(SOMs)可用于表征对象的变化性。为了为基于任务的IQ分析建立现实的SOMs,使用实验图像数据是理想的。然而,从医疗成像系统获得的实验图像数据受到测量噪声的影响。先前的工作研究了使用增强生成对抗网络(GAN)的深生成模型(DGM)的能力,即AmbientGAN,用于从带噪声的测量图像数据中建立SOMs。最近,降噪扩散模型(DDM)已成为领先的图像合成DGM,并且能产生比GAN更高的图像质量。然而,原始DDM具有缓慢的图像处理过程,这是由于去噪步骤中的高斯假设。最近,提出了降噪扩散GAN(DDGAN),以实现快速的图像生成并保持与原始DDM相当的高生成图像质量。在这项工作中,我们提出了一种增强的DDGAN架构,即Ambient DDGAN(ADDGAN),用于从带噪声的图像数据中学习SOMs。我们进行了涉及临床计算机断层扫描(CT)图像和数字乳腺断层合成(DBT)图像的数值研究。所展示的ADDGAN从带噪声的图像数据中学习现实的SOMs的能力得到了验证。结果表明,ADDGAN在合成具有复杂纹理的高分辨率医学图像方面显著优于先进的AmbientGAN模型。
论文及项目相关链接
PDF SPIE Medical Imaging 2025
Summary
本文探讨了使用深度生成模型(DGM)建立基于任务图像质量(IQ)分析的随机对象模型(SOM)的方法。特别关注了新兴的去噪扩散模型(DDMs)及其变体DDGAN和增强的DDGAN架构(ADDGAN)。研究表明,ADDGAN能够从含噪声的图像数据中学习现实的SOM,并在合成具有复杂纹理的高分辨率医学图像方面显著优于先进的AmbientGAN模型。
Key Takeaways
- 医学成像系统的客观评估应基于任务图像质量(IQ)措施,考虑图像数据中的所有随机性来源。
- 随机对象模型(SOMs)可用于表征对象变异性。
- 实验图像数据可用于建立现实的SOMs进行任务IQ分析,但这些数据通常受到测量噪声的影响。
- 深度生成模型(DGM)如去噪扩散模型(DDMs)和DDGAN可用于从含噪声的图像数据中建立SOM。
- 最新提出的增强的DDGAN架构(ADDGAN)可实现快速图像生成,同时保持与原始DDMs相当的高质量图像生成能力。
点此查看论文截图



RTFAST-Spectra: Emulation of X-ray reverberation mapping for active galactic nuclei
Authors:Benjamin Ricketts, Daniela Huppenkothen, Matteo Lucchini, Adam Ingram, Guglielmo Mastroserio, Matthew Ho, Benjamin Wandelt
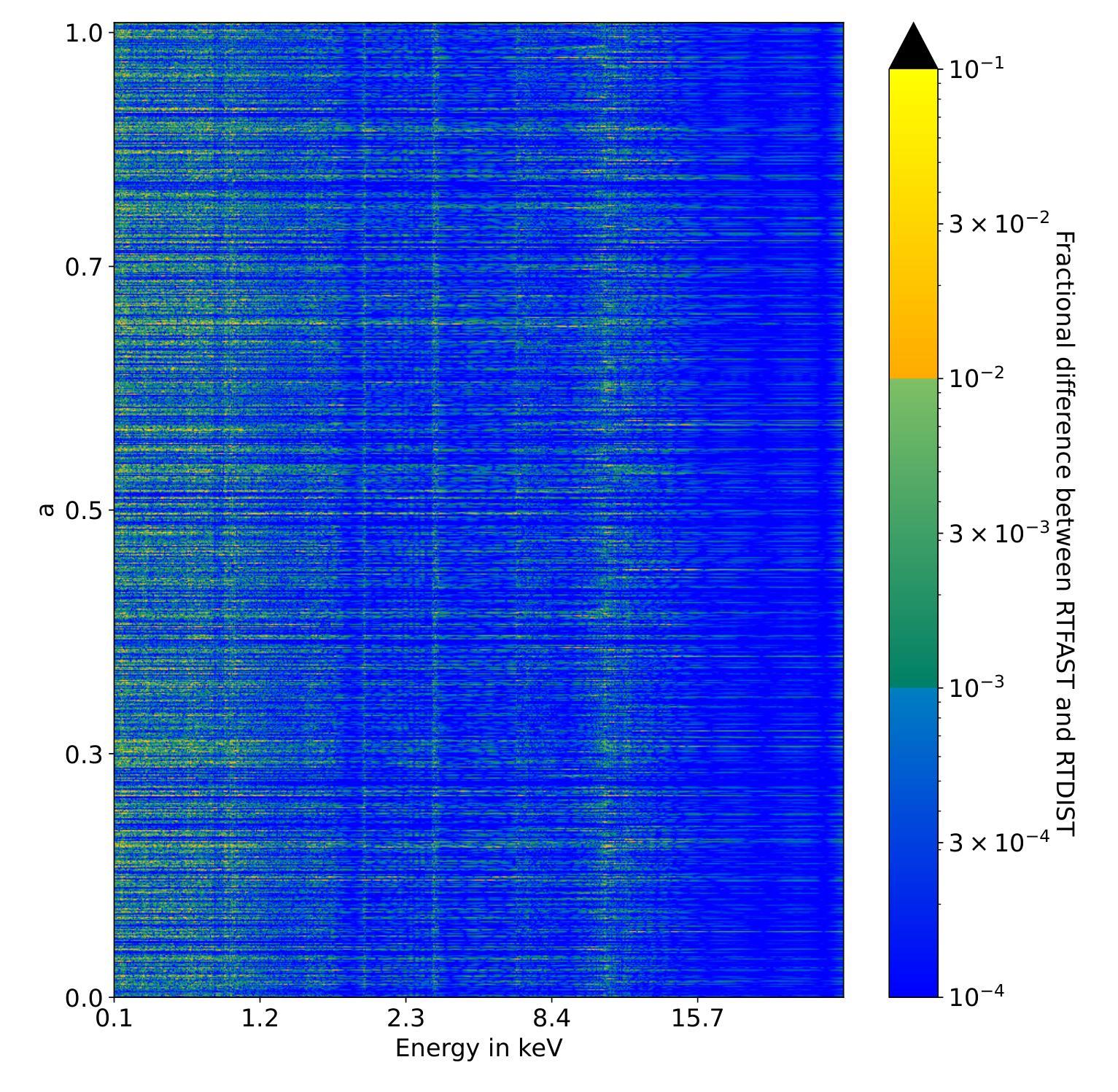
Bayesian analysis has begun to be more widely adopted in X-ray spectroscopy, but it has largely been constrained to relatively simple physical models due to limitations in X-ray modelling software and computation time. As a result, Bayesian analysis of numerical models with high physics complexity have remained out of reach. This is a challenge, for example when modelling the X-ray emission of accreting black hole X-ray binaries, where the slow model computations severely limit explorations of parameter space and may bias the inference of astrophysical parameters. Here, we present RTFAST-Spectra: a neural network emulator that acts as a drop in replacement for the spectral portion of the black hole X-ray reverberation model RTDIST. This is the first emulator for the reltrans model suite and the first emulator for a state-of-the-art x-ray reflection model incorporating relativistic effects with 17 physically meaningful model parameters. We use Principal Component Analysis to create a light-weight neural network that is able to preserve correlations between complex atomic lines and simple continuum, enabling consistent modelling of key parameters of scientific interest. We achieve a $\mathcal{O}(10^2)$ times speed up over the original model in the most conservative conditions with $\mathcal{O}(1%)$ precision over all 17 free parameters in the original numerical model, taking full posterior fits from months to hours. We employ Markov Chain Monte Carlo sampling to show how we can better explore the posteriors of model parameters in simulated data and discuss the complexities in interpreting the model when fitting real data.
贝叶斯分析在X射线光谱学中已经开始得到更广泛的应用,但由于X射线建模软件和计算时间的限制,它主要局限于相对简单的物理模型。因此,具有复杂物理数值模型的贝叶斯分析仍然无法达到。当对吸积黑洞X射线双星的X射线发射进行建模时,这是一个挑战。慢速模型计算严重限制了参数空间的探索,并可能导致天体物理参数的推断出现偏差。在这里,我们介绍了RTFAST-Spectra:一个神经网络模拟器,它可以作为黑洞X射线回波模型RTDIST光谱部分的替代。这是第一个为reltrans模型套件和第一个为最新x射线反射模型设计的模拟器,该模型结合了相对论效应,具有17个物理意义明确的模型参数。我们使用主成分分析创建了一个轻量级的神经网络,能够保持复杂原子线和简单连续体之间的相关性,从而能够对科学兴趣的关键参数进行一致建模。我们在最保守的条件下实现了相对于原始模型的$\mathcal{O}(10^2)$倍加速,在原始模型的全部17个自由参数上实现了$\mathcal{O}(1%)$的精确度,将全后验拟合从数月缩短至数小时。我们采用马尔可夫链蒙特卡罗采样法,展示了如何更好地探索模拟数据的模型参数后验分布,并讨论了在实际数据拟合中解释模型的复杂性。
论文及项目相关链接
PDF 22 pages, 35 figures. Accepted in MNRAS
摘要
本文介绍了RTFAST-Spectra神经网络模拟器在X射线光谱分析中的应用。该模拟器可作为黑洞X射线回波模型RTDIST光谱部分的替代方案。它是首个为reltrans模型套件和首个为一流的考虑相对论效应的X射线反射模型创建的模拟器,该模型包含17个具有物理意义的模型参数。使用主成分分析创建轻量级神经网络,能够保留复杂原子线和简单连续谱之间的相关性,实现对关键科学参数的一致建模。在最保守的条件下,相对于原始模型实现了$\mathcal{O}(10^2)$倍的速度提升,在所有的17个自由参数上保持$\mathcal{O}(1%)$的精度。采用马尔可夫链蒙特卡罗采样,展示了如何更好地探索模拟数据的模型参数后验分布,并讨论了拟合真实数据时模型解释的复杂性。
关键见解
- Bayesian分析在X射线光谱中的应用受到软件和计算时间的限制,难以处理高物理复杂度的数值模型。
- RTFAST-Spectra是首个为reltrans模型套件和考虑相对论效应的先进X射线反射模型创建的模拟器。
- 使用主成分分析创建轻量级神经网络,能够保留复杂原子线和简单连续谱之间的相关性。
- 实现了对原始模型$\mathcal{O}(10^2)$倍的速度提升,同时在所有17个自由参数上保持$\mathcal{O}(1%)$的精度。
- 通过Markov Chain Monte Carlo采样,更好地探索了模拟数据中的模型参数后验分布。
- 在拟合真实数据时,模型解释的复杂性被讨论。
- 该模拟器有望改善X射线光谱分析中的参数空间探索,减少计算时间,促进Bayesian分析在复杂物理模型中的应用。
点此查看论文截图

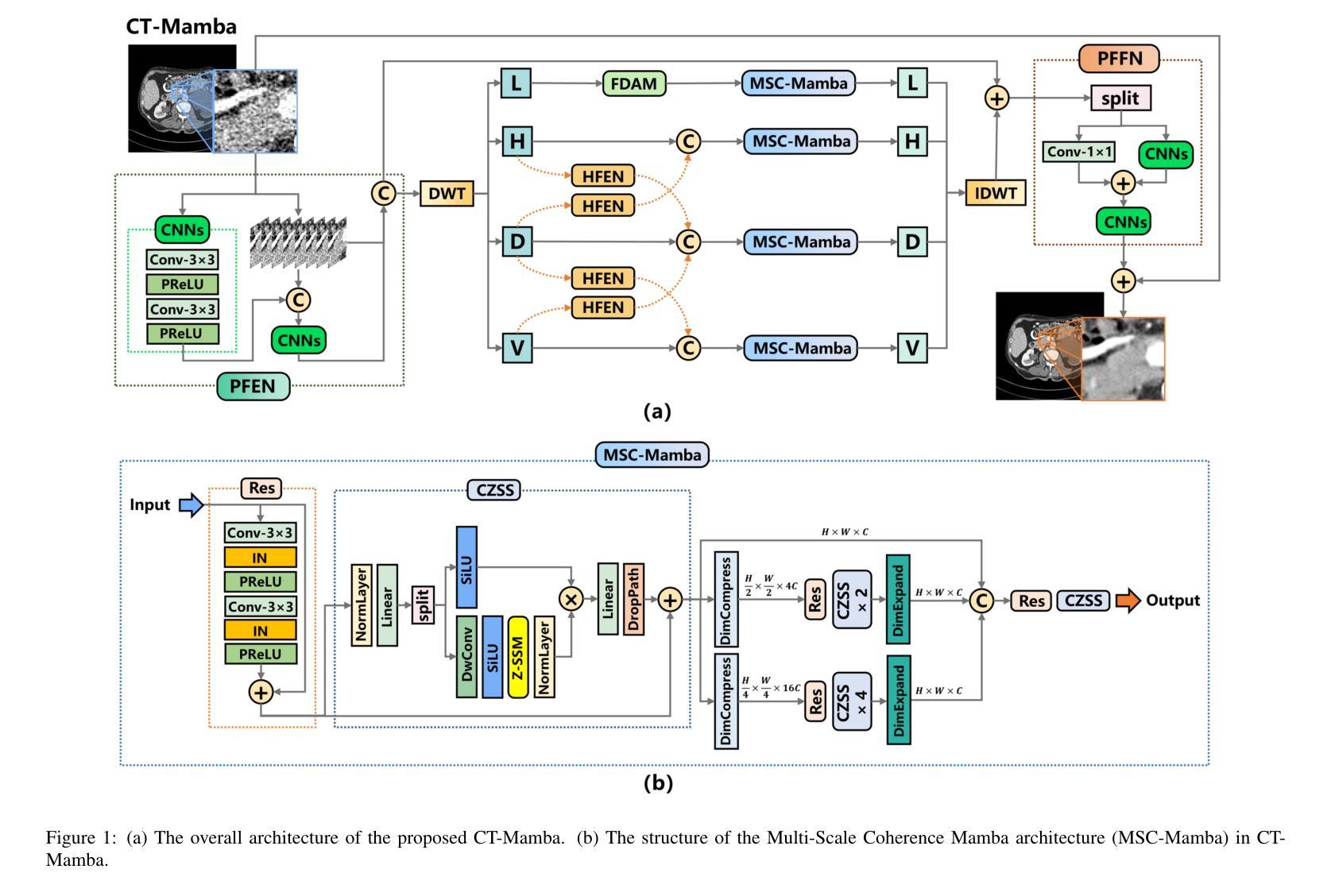
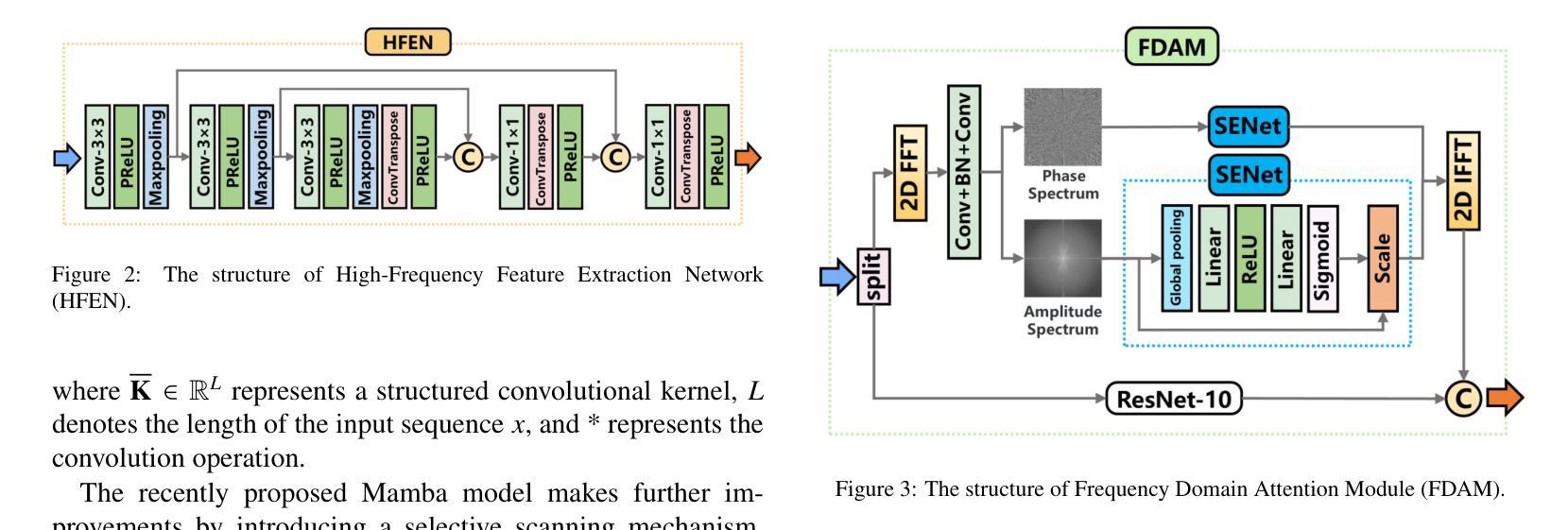
CT-Mamba: A Hybrid Convolutional State Space Model for Low-Dose CT Denoising
Authors:Linxuan Li, Wenjia Wei, Luyao Yang, Wenwen Zhang, Jiashu Dong, Yahua Liu, Wei Zhao
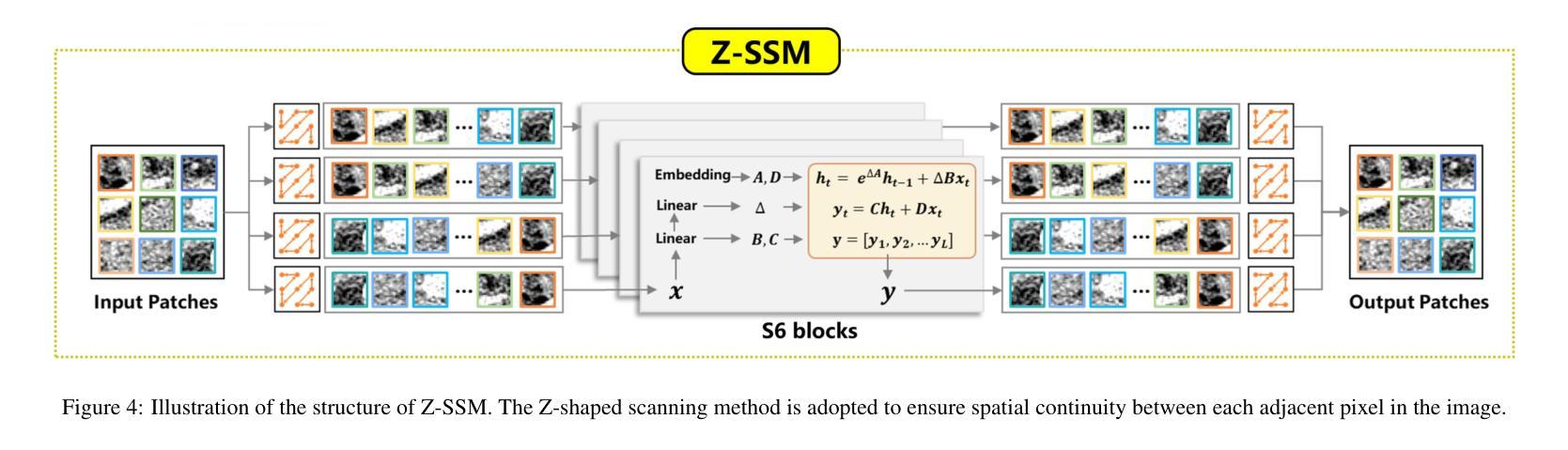
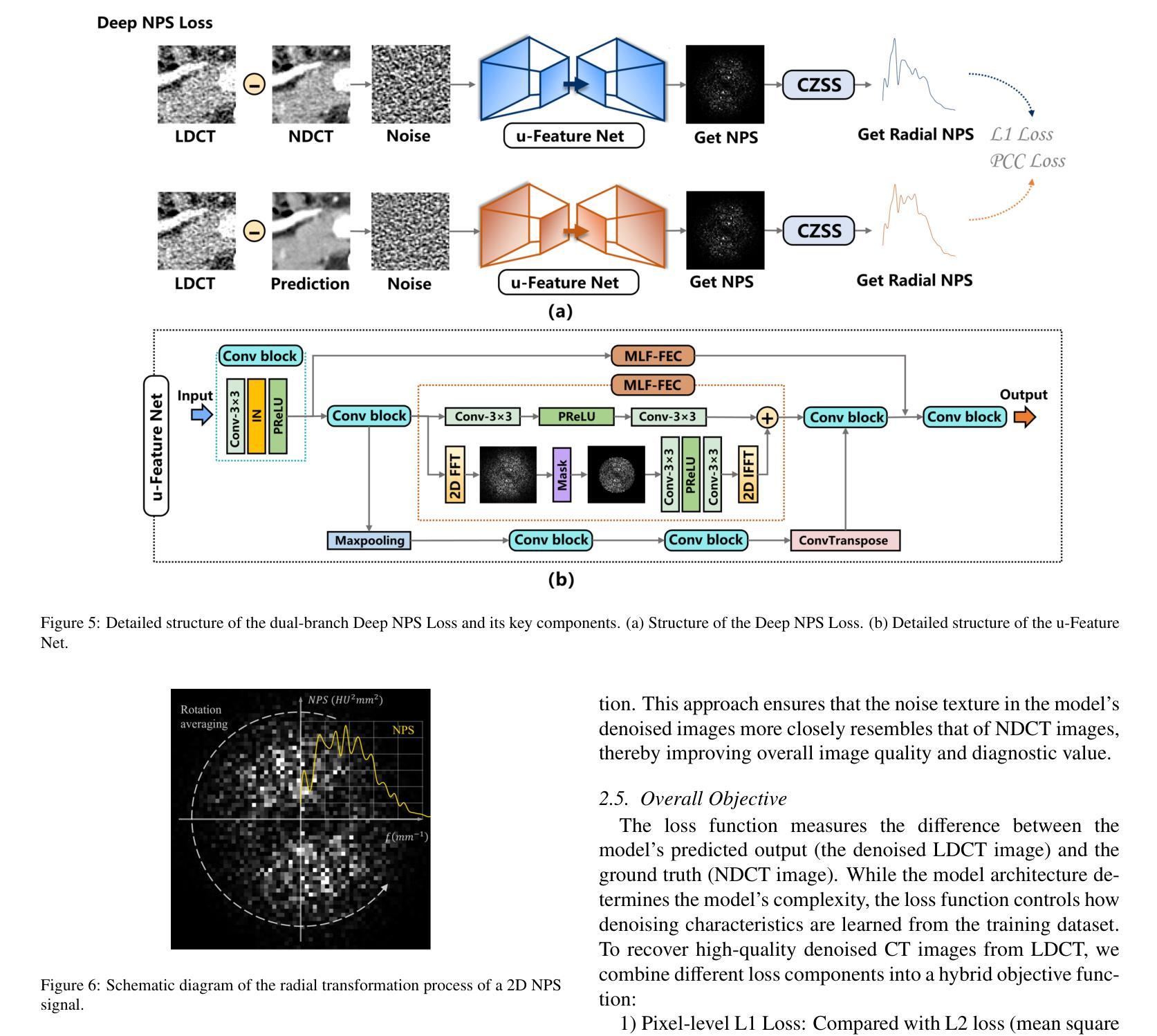
Low-dose CT (LDCT) significantly reduces the radiation dose received by patients, however, dose reduction introduces additional noise and artifacts. Currently, denoising methods based on convolutional neural networks (CNNs) face limitations in long-range modeling capabilities, while Transformer-based denoising methods, although capable of powerful long-range modeling, suffer from high computational complexity. Furthermore, the denoised images predicted by deep learning-based techniques inevitably exhibit differences in noise distribution compared to normal-dose CT (NDCT) images, which can also impact the final image quality and diagnostic outcomes. This paper proposes CT-Mamba, a hybrid convolutional State Space Model for LDCT image denoising. The model combines the local feature extraction advantages of CNNs with Mamba’s strength in capturing long-range dependencies, enabling it to capture both local details and global context. Additionally, we introduce an innovative spatially coherent ‘Z’-shaped scanning scheme to ensure spatial continuity between adjacent pixels in the image. We design a Mamba-driven deep noise power spectrum (NPS) loss function to guide model training, ensuring that the noise texture of the denoised LDCT images closely resembles that of NDCT images, thereby enhancing overall image quality and diagnostic value. Experimental results have demonstrated that CT-Mamba performs excellently in reducing noise in LDCT images, enhancing detail preservation, and optimizing noise texture distribution, and exhibits higher statistical similarity with the radiomics features of NDCT images. The proposed CT-Mamba demonstrates outstanding performance in LDCT denoising and holds promise as a representative approach for applying the Mamba framework to LDCT denoising tasks. Our code will be made available after the paper is officially published: https://github.com/zy2219105/CT-Mamba/.
低剂量CT(LDCT)显著减少了患者接受的辐射剂量,然而,剂量的减少会引入额外的噪声和伪影。目前,基于卷积神经网络(CNNs)的降噪方法在长期建模能力方面存在局限性,而基于Transformer的降噪方法虽然具有强大的长期建模能力,但计算复杂度较高。此外,基于深度学习技术预测的降噪图像与正常剂量CT(NDCT)图像相比,噪声分布不可避免地存在差异,这也可能影响最终的图像质量和诊断结果。本文提出了CT-Mamba,一种用于LDCT图像降噪的混合卷积状态空间模型。该模型结合了CNNs提取局部特征的优势和Mamba在捕捉长期依赖关系方面的实力,使其能够捕捉局部细节和全局上下文。此外,我们引入了一种创新的空间连贯性“Z”形扫描方案,以确保图像中相邻像素之间的空间连续性。我们设计了一个由Mamba驱动的深度噪声功率谱(NPS)损失函数来引导模型训练,确保降噪LDCT图像的噪声纹理与NDCT图像相似,从而提高整体图像质量和诊断价值。实验结果表明,CT-Mamba在降低LDCT图像噪声、提高细节保留和优化噪声纹理分布方面表现出色,与NDCT图像的放射学特征具有更高的统计相似性。所提出的CT-Mamba在LDCT降噪方面表现出卓越的性能,并有望作为将Mamba框架应用于LDCT降噪任务的一种代表性方法。我们的代码将在论文正式发表后提供:https://github.com/zy2219105/CT-Mamba/。
论文及项目相关链接
摘要
这篇论文提出一种用于低剂量CT(LDCT)图像去噪的混合卷积状态空间模型——CT-Mamba。该模型结合了卷积神经网络(CNNs)的局部特征提取优势和Mamba在捕捉长距离依赖方面的优势,既能捕捉局部细节又能捕捉全局上下文。此外,还引入了一种创新的Z形扫描方案,确保图像中相邻像素之间的空间连续性。设计了一种基于Mamba的深度噪声功率谱(NPS)损失函数,以指导模型训练,确保去噪后的LDCT图像的噪声纹理与常规剂量CT(NDCT)图像相似,从而提高整体图像质量和诊断价值。实验结果表明,CT-Mamba在降低LDCT图像噪声、增强细节保留和优化噪声纹理分布方面表现出卓越的性能,与NDCT图像的放射学特征表现出较高的统计相似性。
关键见解
- 低剂量CT(LDCT)能够显著减少患者接受的辐射剂量,但剂量减少会导致额外的噪声和伪影。
- 当前基于卷积神经网络(CNNs)的去噪方法在长期建模能力方面存在局限性,而基于Transformer的去噪方法虽然具有强大的长期建模能力,但计算复杂度较高。
- 深度学习技术预测的降噪图像与常规剂量CT(NDCT)图像的噪声分布存在差异,可能影响最终图像质量和诊断结果。
- CT-Mamba模型结合了CNN和Mamba的优势,能够捕捉局部细节和全局上下文。
- 创新的Z形扫描方案确保图像中相邻像素之间的空间连续性。
- 引入深度噪声功率谱(NPS)损失函数,以指导模型训练,使去噪后的LDCT图像的噪声纹理更接近NDCT图像。
- 实验结果表明,CT-Mamba在降低LDCT图像噪声、提高细节保留和噪声纹理分布优化方面表现出卓越性能。
点此查看论文截图

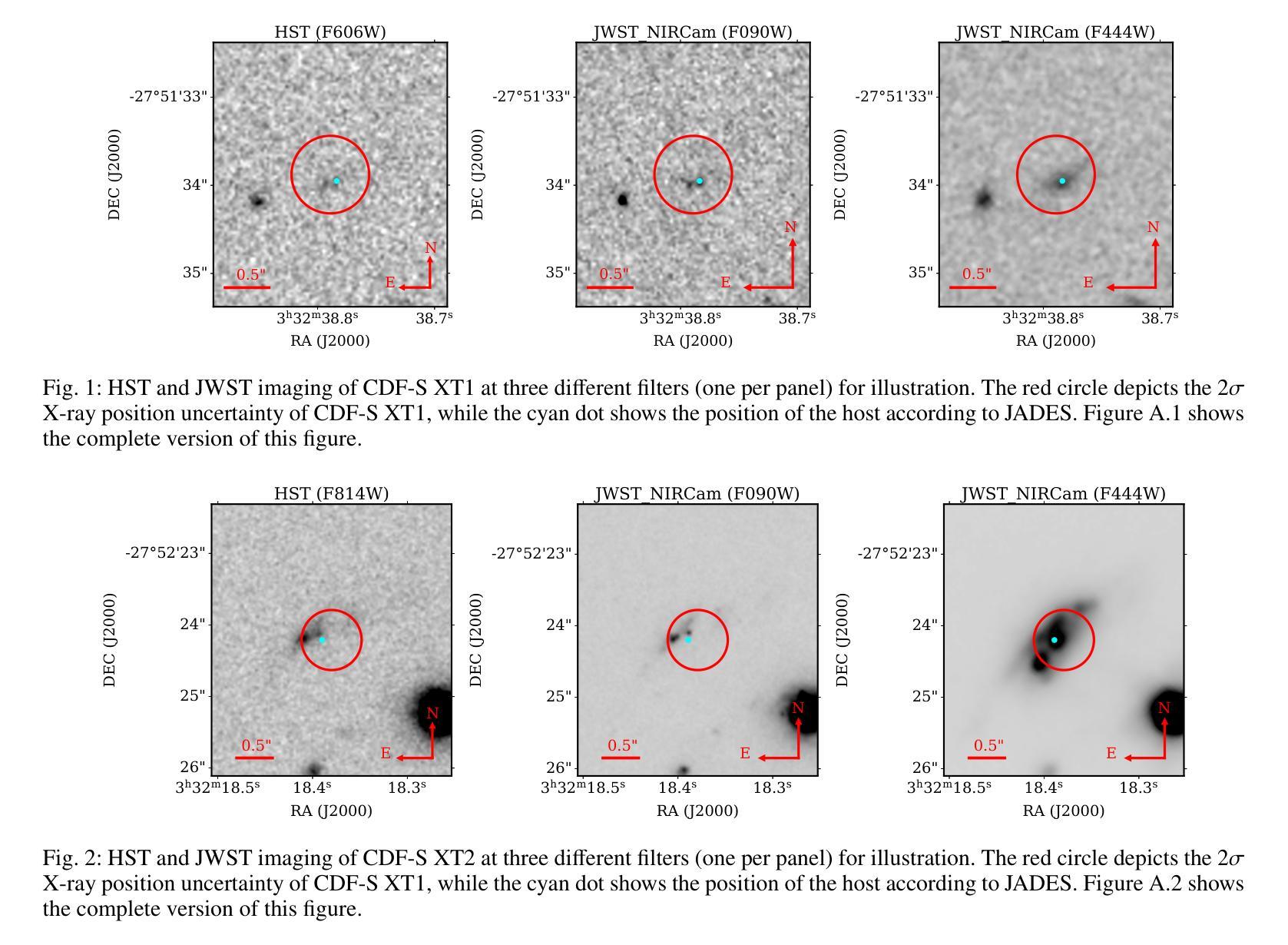
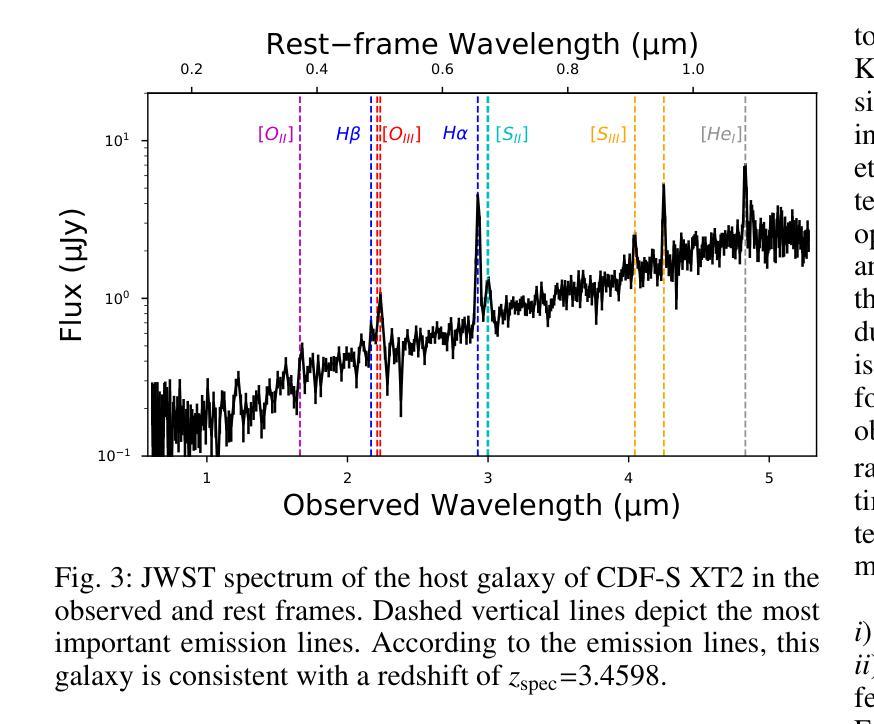
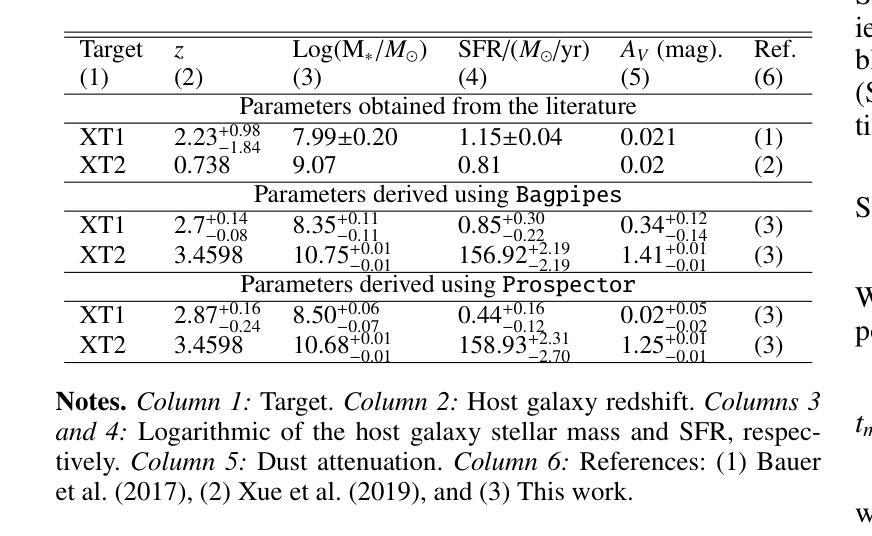
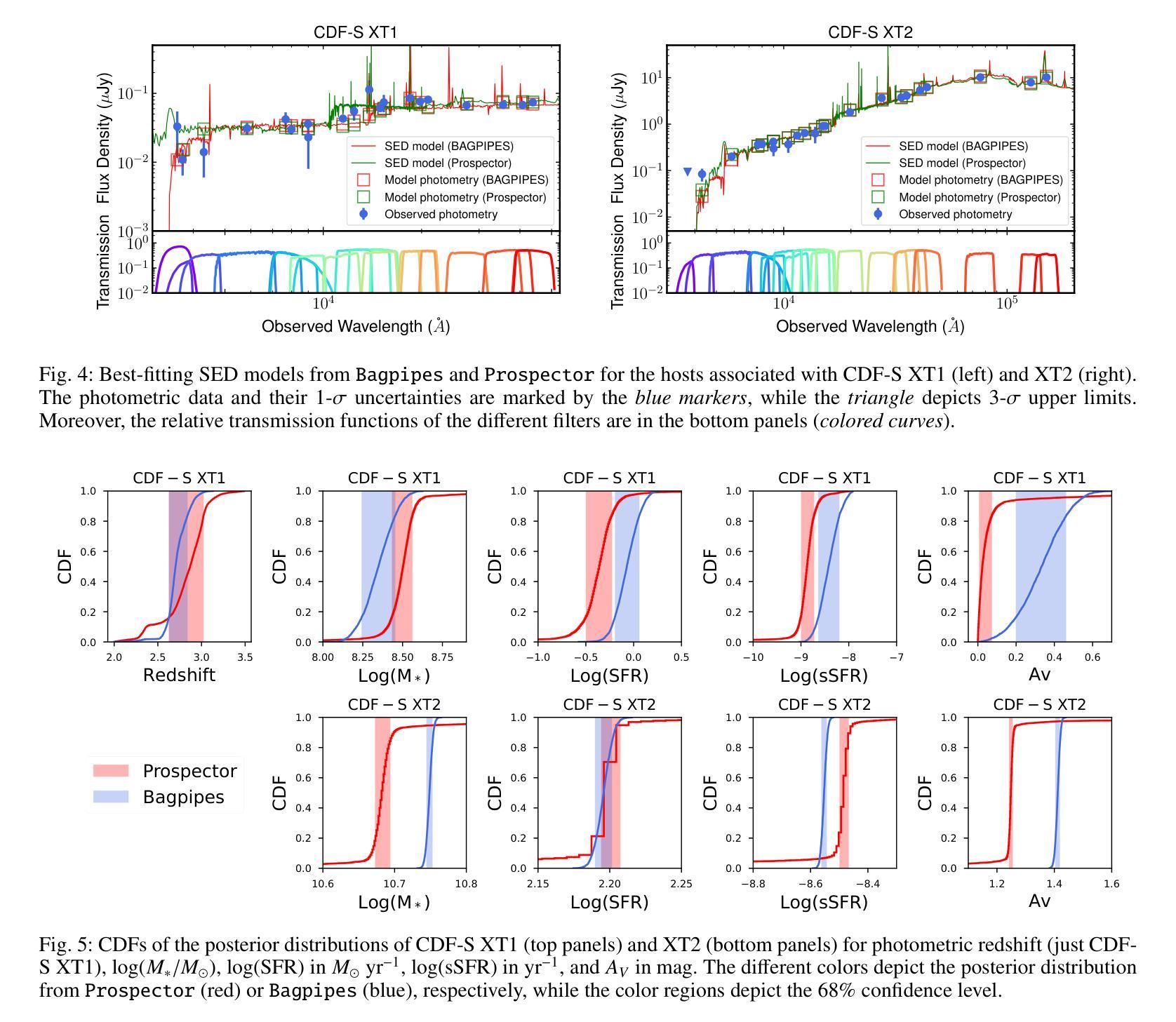
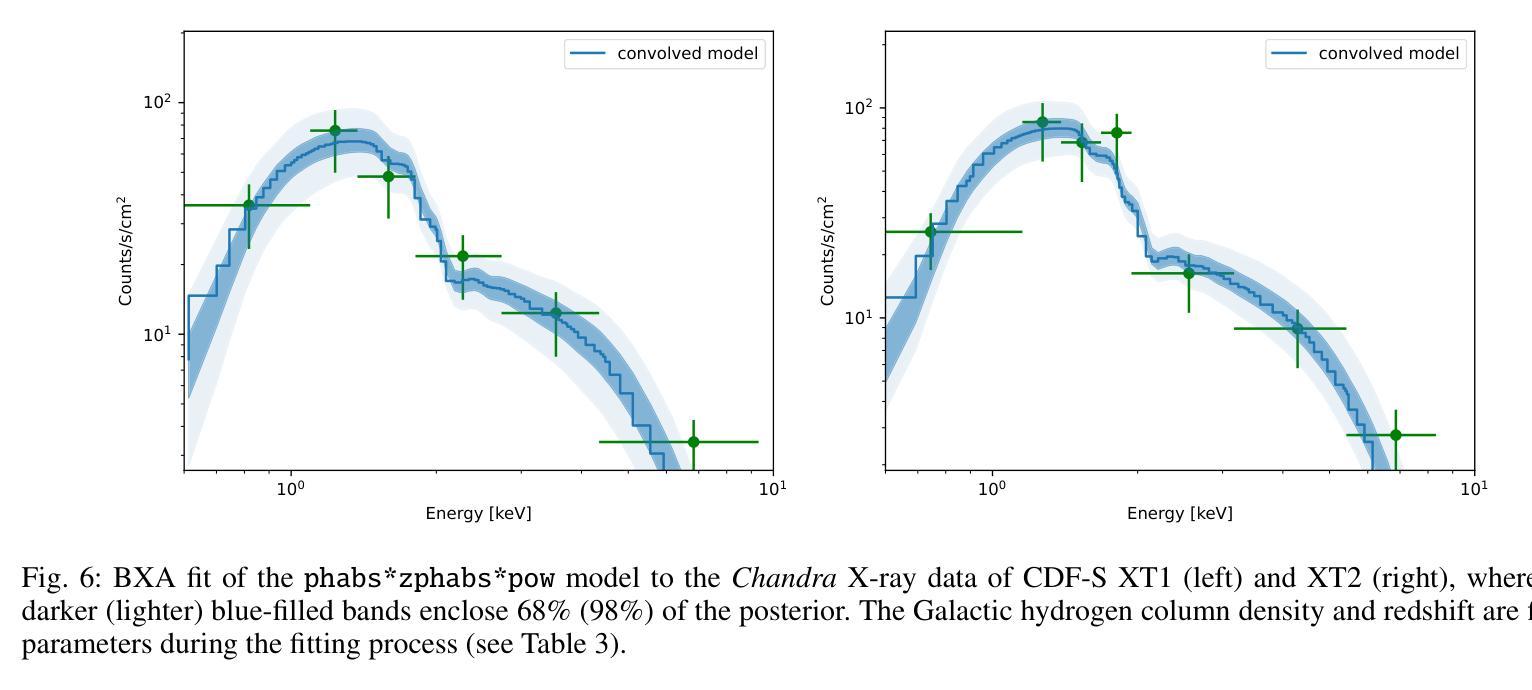
New JWST redshifts for the host galaxies of CDF-S XT1 and XT2: understanding their nature
Authors:J. Quirola-Vásquez, F. E. Bauer, P. G. Jonker, A. Levan, W. N. Brandt, M. Ravasio, D. Eappachen, Y. Q. Xue, X. C. Zheng
CDF-S XT1 and XT2 are considered two canonical extragalactic fast X-ray transients (FXTs). In this work, we report new constraints on both FXTs, based on recent JWST NIRCam and MIRI photometry, as well as NIRspec spectroscopy for CDF-S XT2 that allow us to improve our understanding of their distances, energetics, and host galaxy properties compared to the pre-JWST era. We use the available HST and JWST archival data to determine the host properties and constrain the energetics of each FXT based on spectral energy distribution (SED) photometric fitting. The host of CDF-S XT1 is now constrained to lie at $z$=2.76, implying a host absolute magnitude $M_R=-19.14$mag, stellar mass $M_{*}=$1.8e8$M_\odot$, and star formation rate SFR$=0.62 M_\odot$/yr. These properties lie at the upper end of previous estimates, leaving CDF-S XT1 with a peak X-ray luminosity of 2.8e47 erg/s. We argue that the best progenitor scenario for XT1 is a low-luminosity gamma-ray burst (GRB), although we do not fully rule out a proto-magnetar association or a jetted tidal disruption event involving a white dwarf and an intermediate-massive black hole. In the case of CDF-S XT2, JWST imaging reveals a new highly obscured component of the host galaxy, previously missed in HST images, while NIRspec spectroscopy securely places the host at $z$=3.4598. The new redshift implies a host with $M_R=-21.76$mag, $M_*=5.5e10 M_\odot$, SFR=160$M_\odot$/yr, and FXT $L_{X,peak}=1.4e47$~erg/s. The revised energetics, similarity to X-ray flash event light curves, small host offset, and high host SFR favor a low-luminosity collapsar progenitor for CDF-S XT2. Although a magnetar model is not ruled out, it appears improbable. While these HST and JWST observations shed light on the host galaxies of XT1 and XT2, and by extension, on the nature of FXTs, a unique explanation for both sources remains elusive.
CDF-S XT1和XT2被认为是两种典型的系外快速X射线瞬变(FXTs)。在这项工作中,我们基于最新的JWST NIRCam和MIRI光度法以及CDF-S XT2的NIRspec光谱法,报告了这两种FXTs的新约束。与之前的时代相比,这使我们能够改进对它们的距离、能量和宿主星系性质的理解。我们使用可用的HST和JWST存档数据来确定宿主属性和基于光谱能量分布(SED)光度拟合的每个FXT的能量约束。CDF-S XT1的宿主现在被限制在$z$=2.76的位置,这意味着宿主绝对星等$M_R=-19.14$ mag,恒星质量$M_{}$=1.8e8 $M_\odot$,星形成率SFR=0.62 $M_\odot$/yr。这些属性处于先前估计值的较高端,使CDF-S XT1的峰值X射线光度为2.8e47 erg/s。我们认为XT1的最佳前身星情景是低光度的伽马射线爆发(GRB),尽管我们并不完全排除原磁星关联或涉及白矮星和中等质量黑洞的喷射潮汐撕裂事件。对于CDF-S XT2来说,JWST成像揭示了一个宿主星系中新的高度遮蔽的组件,这在之前的HST图像中未被发现,而NIRspec光谱法将其稳定地定位在$z$=3.4598的位置。新的红移意味着宿主具有$M_R=-21.76$ mag,$M_=5.5e10 M_\odot$,SFR=160 $M_\odot$/yr和FXT $L_{X,peak}=1.4e47$ erg/s。修订后的能量学、与X射线闪光事件光曲线的相似性、较小的宿主偏移以及较高的宿主SFR支持CDF-S XT2为低光度塌星起源的说法。虽然磁星模型并没有被排除,但似乎不太可能。虽然这些HST和JWST观测揭示了XT1和XT2的宿主星系,并由此扩展到了FXTs的性质,但对于两个来源的独特解释仍然难以捉摸。
论文及项目相关链接
PDF The manuscript was accepted by Astronomy & Astrophysics in January 2025
Summary
CDF-S XT1和XT2是两种典型的快速射电星系瞬态事件(FXTs)。利用最新的JWST红外照相机和多模态成像光度的MIRI摄影术以及CDF-S XT2的NIRspec光谱学数据,我们对其距离、能量和宿主星系特性有了更深入的了解。基于光谱能量分布(SED)的光度拟合方法,CDF-S XT1的宿主星系被限制在z=2.76处,其绝对星等、恒星质量和恒星形成率均处于较高的水平。CDF-S XT2的JWST成像揭示了宿主星系中存在的一个新的高度遮蔽组分,NIRspec光谱进一步确认其位于z=3.4598处。尽管对CDF-S XT1的最佳起源假设是低亮度伽马射线爆发(GRB),但我们并不完全排除磁星相关事件或涉及矮星和超质量黑洞的潮汐撕裂事件的可能性。CDF-S XT2更像是一种低亮度超新星事件,与射电星系现象的光曲线相似,但其确切起源仍不明确。这些观测结果为我们理解这两种FXTs的宿主星系提供了关键依据,有助于我们对瞬态天文学有更深入的认识。尽管如此,仍然缺少能够同时解释两个来源的唯一解释。
Key Takeaways
- CDF-S XT1和XT2是两种重要的快速射电星系瞬态事件(FXTs)。
- 利用JWST的最新数据,对CDF-S XT1和XT2的距离、能量和宿主星系特性有了更精确的了解。
- CDF-S XT1可能源自低亮度伽马射线爆发(GRB),但也考虑其他可能性如磁星相关事件或潮汐撕裂事件。
点此查看论文截图

World of Forms: Deformable Geometric Templates for One-Shot Surface Meshing in Coronary CT Angiography
Authors:Rudolf L. M. van Herten, Ioannis Lagogiannis, Jelmer M. Wolterink, Steffen Bruns, Eva R. Meulendijks, Damini Dey, Joris R. de Groot, José P. Henriques, R. Nils Planken, Simone Saitta, Ivana Išgum
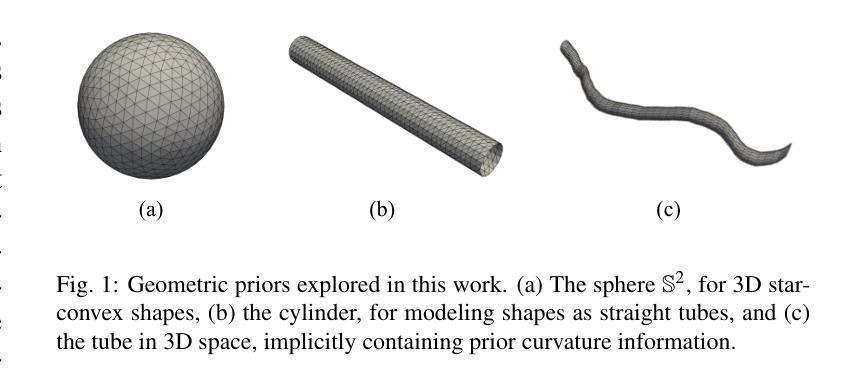
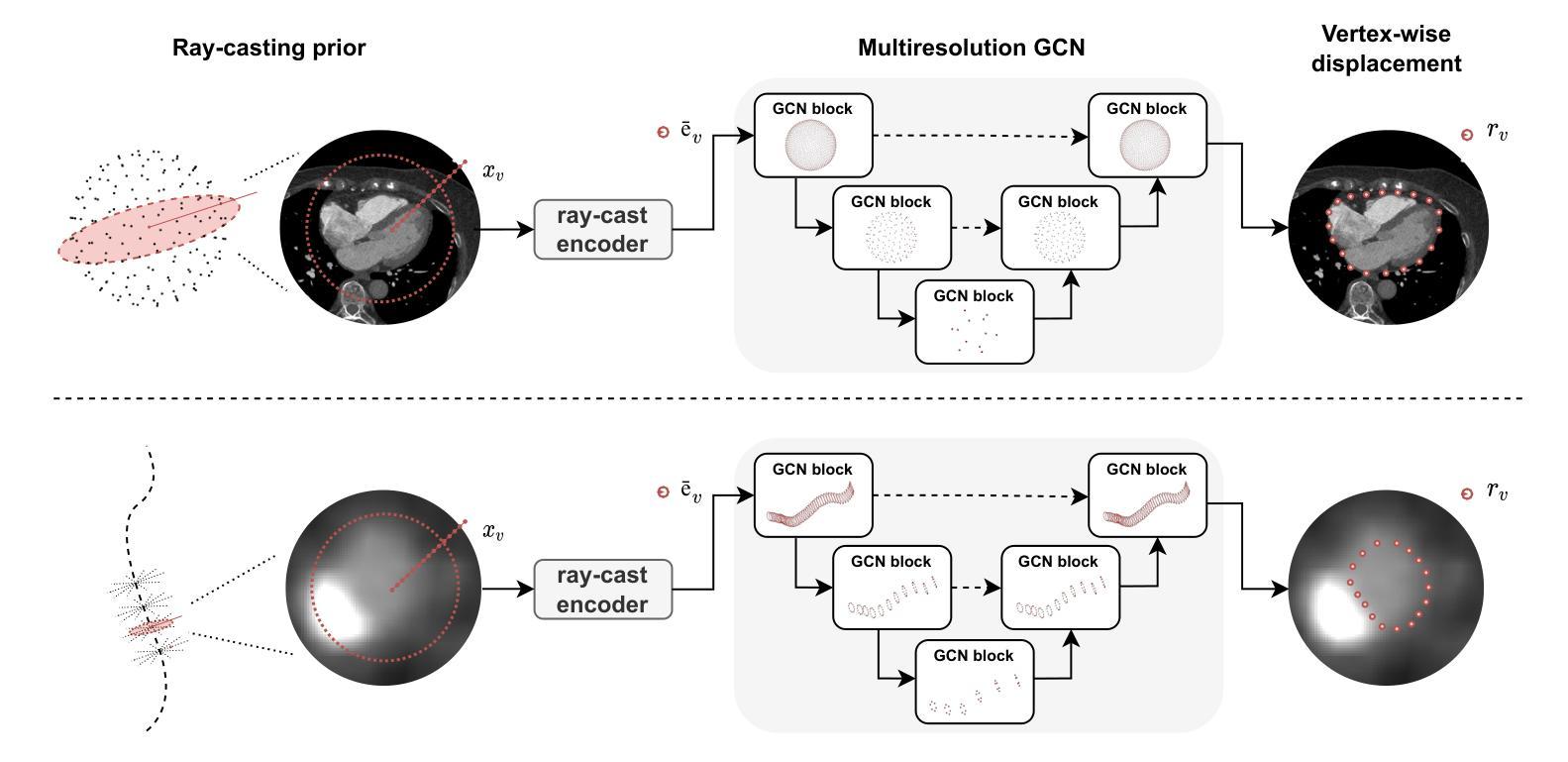
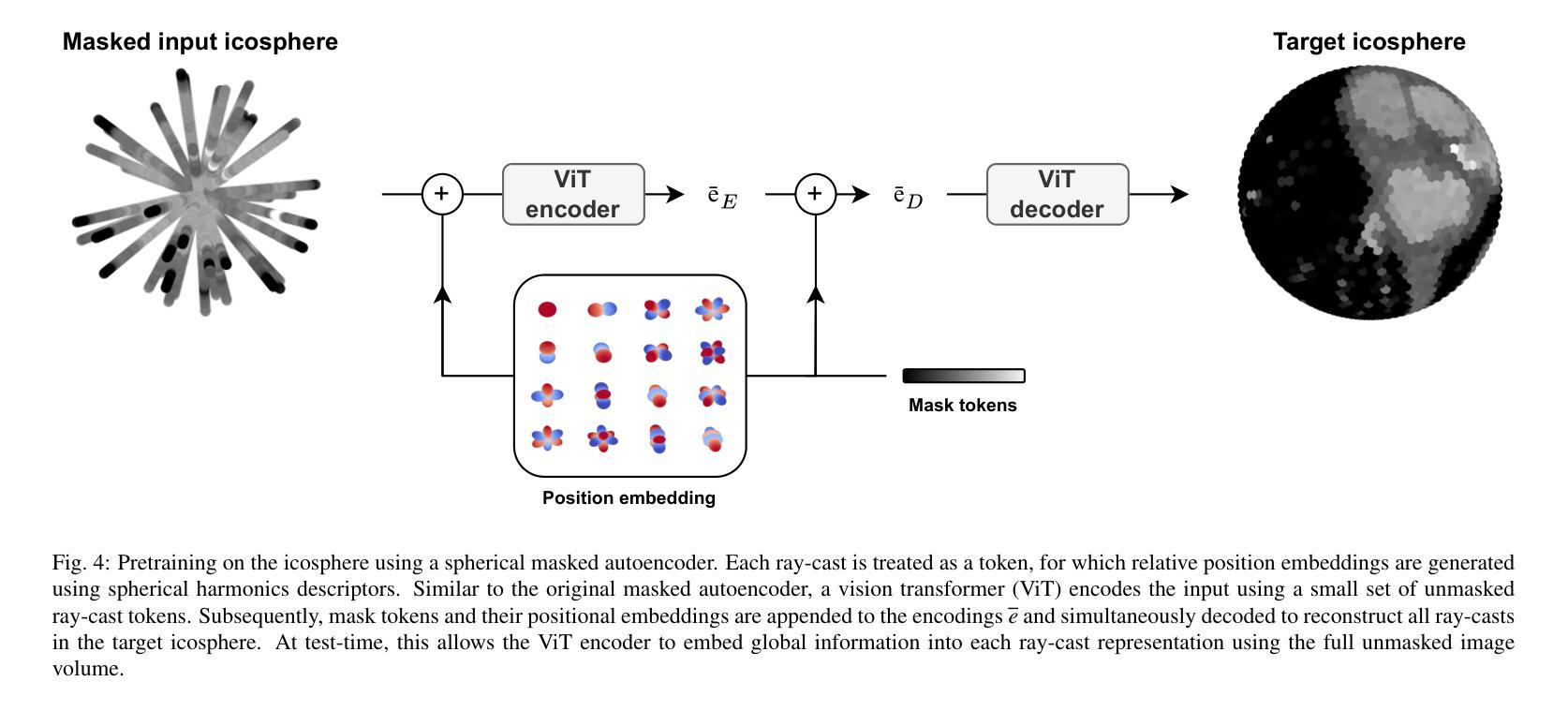
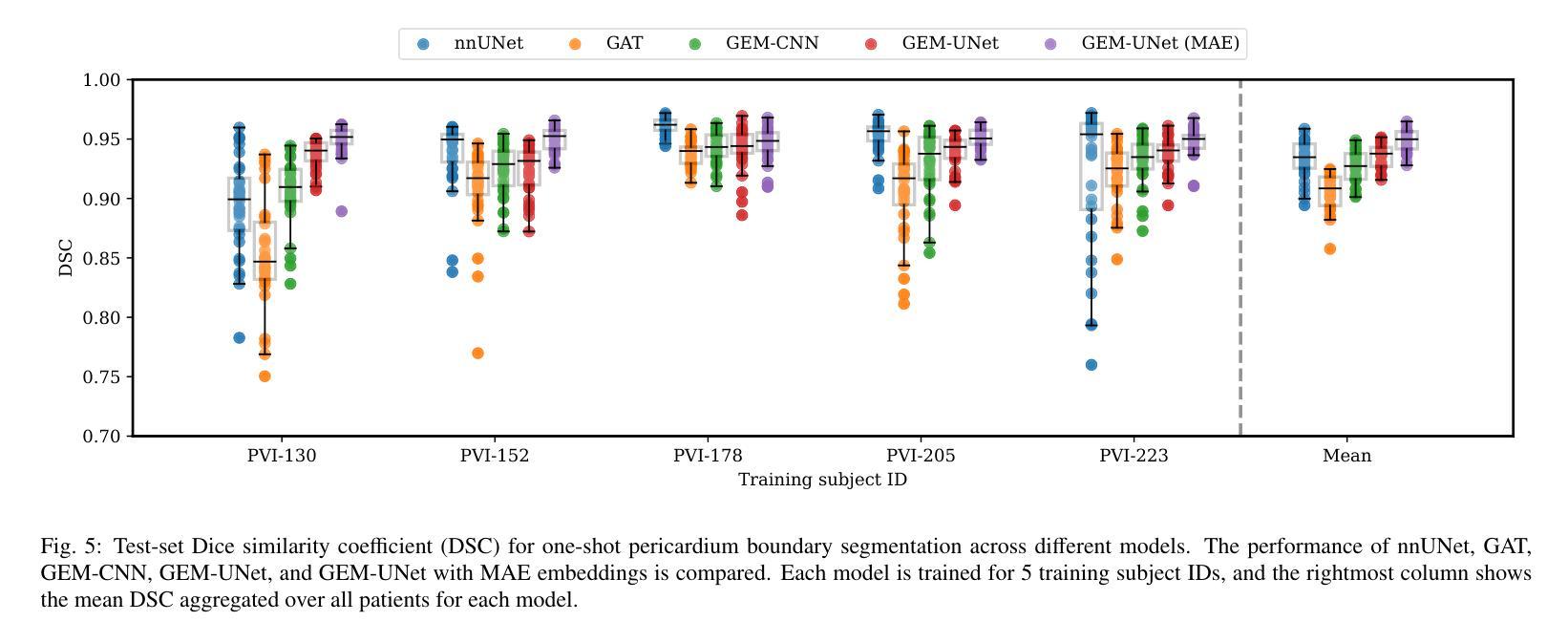
Deep learning-based medical image segmentation and surface mesh generation typically involve a sequential pipeline from image to segmentation to meshes, often requiring large training datasets while making limited use of prior geometric knowledge. This may lead to topological inconsistencies and suboptimal performance in low-data regimes. To address these challenges, we propose a data-efficient deep learning method for direct 3D anatomical object surface meshing using geometric priors. Our approach employs a multi-resolution graph neural network that operates on a prior geometric template which is deformed to fit object boundaries of interest. We show how different templates may be used for the different surface meshing targets, and introduce a novel masked autoencoder pretraining strategy for 3D spherical data. The proposed method outperforms nnUNet in a one-shot setting for segmentation of the pericardium, left ventricle (LV) cavity and the LV myocardium. Similarly, the method outperforms other lumen segmentation operating on multi-planar reformatted images. Results further indicate that mesh quality is on par with or improves upon marching cubes post-processing of voxel mask predictions, while remaining flexible in the choice of mesh triangulation prior, thus paving the way for more accurate and topologically consistent 3D medical object surface meshing.
基于深度学习的医学图像分割和表面网格生成通常涉及从图像到分割再到网格的序列流程,这需要大量的训练数据集,而先验几何知识的利用却很有限。这可能导致拓扑不一致和在低数据环境下的性能不佳。为了解决这些挑战,我们提出了一种利用几何先验进行直接3D解剖对象表面网格化的高效数据深度学习方法。我们的方法采用多分辨率图神经网络,在先验几何模板上运行,该模板会变形以适应目标对象的边界。我们展示了如何为不同的表面网格目标使用不同的模板,并引入了用于3D球形数据的全新掩码自编码器预训练策略。所提出的方法在一次成像设置中对心包、左心室腔和左心室心肌的分割表现优于nnUNet。同样,该方法在针对多平面重建图像的操作中的腔室分割表现也优于其他方法。结果还表明,网格质量可与基于体素掩膜预测的八叉树行进算法的后处理相当或更好,同时在网格三角剖分先验的选择上具有灵活性,从而为更准确和拓扑一致的3D医学对象表面网格化铺平了道路。
论文及项目相关链接
PDF Submitted to Medical Image Analysis
Summary
本文提出一种数据高效的深度学习方法,用于基于几何先验的直接三维解剖对象表面网格生成。该方法采用多分辨率图神经网络,在先验几何模板上进行操作,通过变形以拟合感兴趣对象的边界。该方法解决了在医学图像分割和表面网格生成中面临的拓扑不一致和低数据环境下的性能不佳等问题。
Key Takeaways
- 深度学习在医学图像分割和表面网格生成中面临挑战,如拓扑不一致性和低数据环境下的性能问题。
- 提出一种数据高效的深度学习方法,直接进行三维解剖对象表面网格生成,利用几何先验信息。
- 采用多分辨率图神经网络,操作先验几何模板,通过变形以拟合对象边界。
- 方法可用于不同的表面网格生成目标,使用不同的模板。
- 引入一种新的掩码自动编码器预训练策略,适用于三维球形数据。
- 该方法在单镜头设置下对心包、左心室腔和左心室心肌的分割表现出优于nnUNet的性能。
点此查看论文截图


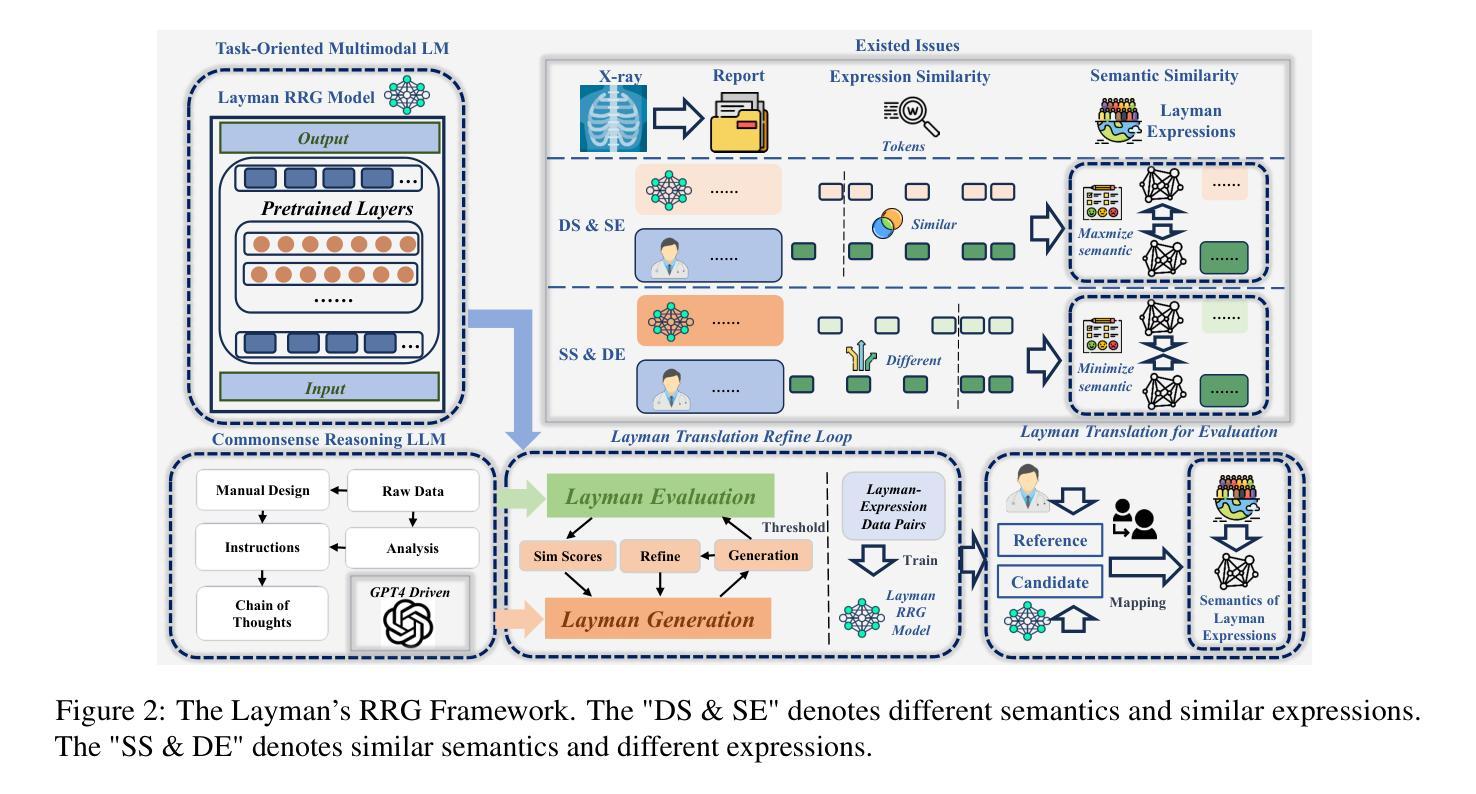
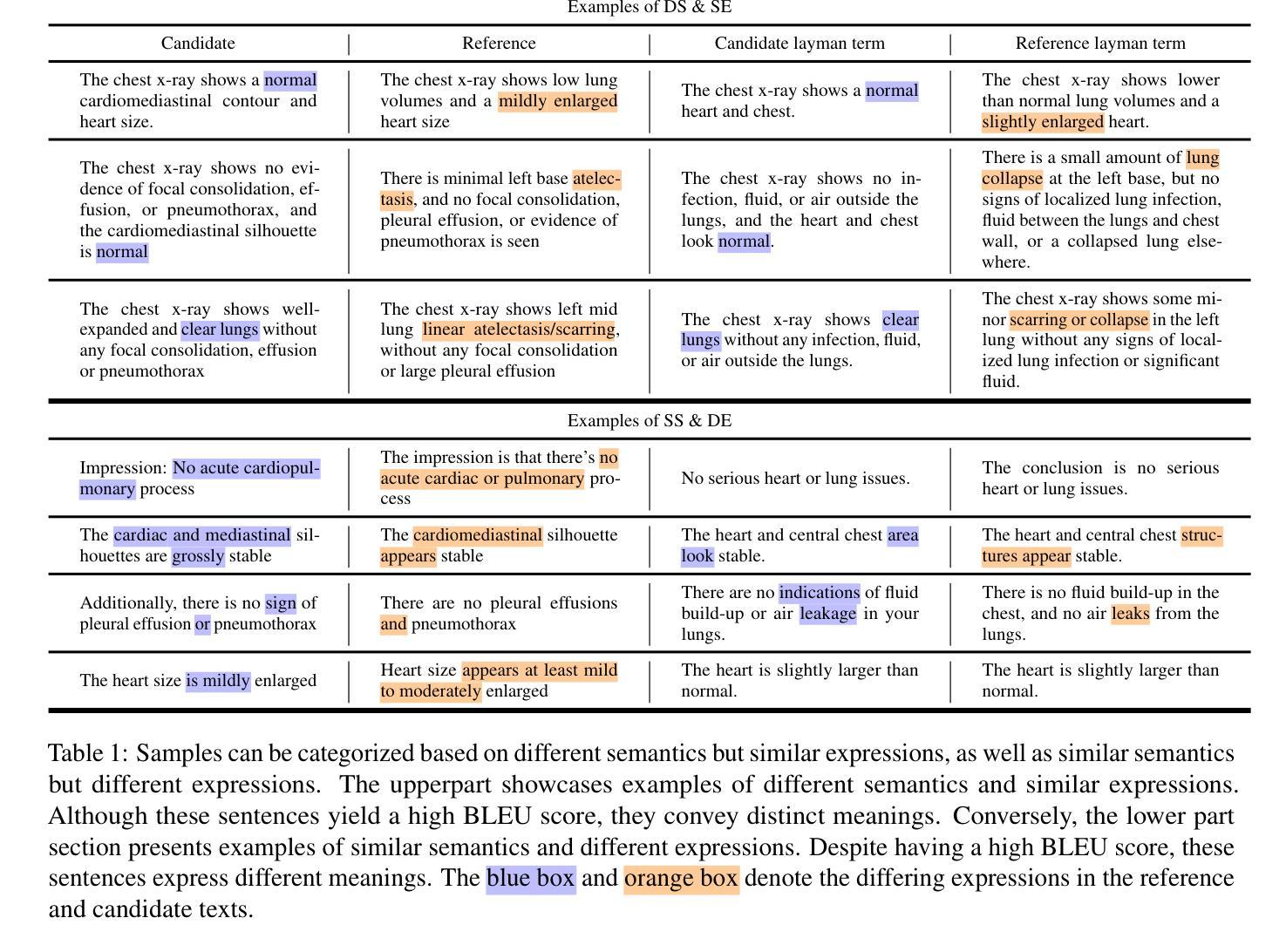
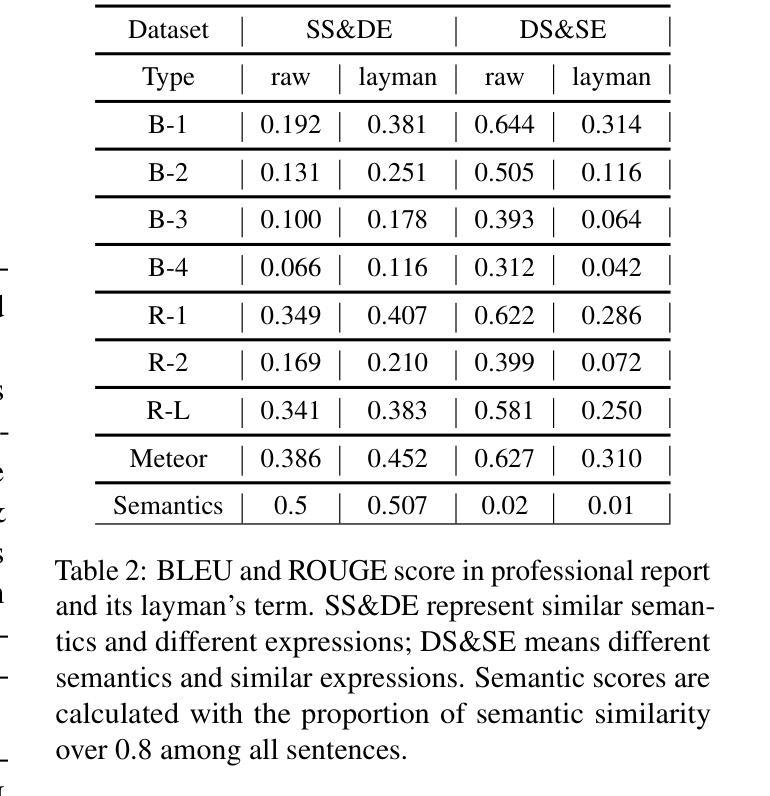
X-ray Made Simple: Radiology Report Generation and Evaluation with Layman’s Terms
Authors:Kun Zhao, Chenghao Xiao, Chen Tang, Bohao Yang, Kai Ye, Noura Al Moubayed, Liang Zhan, Chenghua Lin
Radiology Report Generation (RRG) has achieved significant progress with the advancements of multimodal generative models. However, the evaluation in the domain suffers from a lack of fair and robust metrics. We reveal that, high performance on RRG with existing lexical-based metrics (e.g. BLEU) might be more of a mirage - a model can get a high BLEU only by learning the template of reports. This has become an urgent problem for RRG due to the highly patternized nature of these reports. In this work, we un-intuitively approach this problem by proposing the Layman’s RRG framework, a layman’s terms-based dataset, evaluation and training framework that systematically improves RRG with day-to-day language. We first contribute the translated Layman’s terms dataset. Building upon the dataset, we then propose a semantics-based evaluation method, which is proved to mitigate the inflated numbers of BLEU and provides fairer evaluation. Last, we show that training on the layman’s terms dataset encourages models to focus on the semantics of the reports, as opposed to overfitting to learning the report templates. We reveal a promising scaling law between the number of training examples and semantics gain provided by our dataset, compared to the inverse pattern brought by the original formats. Our code is available at https://github.com/hegehongcha/LaymanRRG.
随着多模态生成模型的进步,放射学报告生成(RRG)已经取得了显著进展。然而,该领域的评估缺乏公平和稳健的指标。我们揭示,在现有的基于词汇的评估指标(例如BLEU)上表现优异的RRG可能更像是一个幻象——模型只需学习报告的模板即可获得高BLEU分数。由于报告的高度模式化特性,这对RRG来说已经成为了一个紧迫的问题。在这项工作中,我们通过提出Layman’s RRG框架来非传统地解决此问题。这是一个基于外行术语的数据集、评估和培训框架,它系统地使用日常语言改进了RRG。我们首先贡献翻译后的外行术语数据集。在此基础上,我们提出了一种基于语义的评估方法,该方法被证明可以缓解BLEU的膨胀数字,并提供更公平的评估。最后,我们展示在普通术语数据集上进行训练会鼓励模型关注报告的语义,而不是过度拟合学习报告模板。我们揭示了与原始格式带来的逆向模式相比,我们的数据集提供的训练示例数量和语义增益之间的一个令人鼓舞的规模化规律。我们的代码位于:https://github.com/hegehongcha/LaymanRRG。
论文及项目相关链接
PDF This paper has substantial data and conceptual changes since release that go beyond simple updating the existing one. As a result, the authors have changed and we need to re-coordinate and reach consensus. So we decide to withdraw it
Summary
随着多模态生成模型的发展,放射学报告生成(RRG)已经取得了显著进步。然而,该领域的评估缺乏公平和稳健的指标。现有基于词汇的评估指标(如BLEU)可能存在误导性,模型只需学习报告模板即可获得高分。针对这一问题,本文提出了平民化RRG框架,该框架基于平民术语数据集、评估和训练框架,系统地改进了RRG的日常语言使用。本文首先贡献翻译后的平民术语数据集,然后在此基础上提出了基于语义的评估方法,该方法缓解了BLEU指标的膨胀数字问题,提供了更公平的评估。最后,本文展示了在平民术语数据集上训练模型能够鼓励模型关注报告的语义内容,而不是过度拟合报告模板。
Key Takeaways
- 多模态生成模型在放射学报告生成(RRG)方面取得显著进展,但评估指标缺乏公平性和稳健性。
- 现有基于词汇的评估指标(如BLEU)可能误导,模型只需学习报告模板即可获得高分。
- 提出平民化RRG框架,使用日常语言,改进RRG的系统性。
- 贡献翻译后的平民术语数据集,为RRG提供新的资源。
- 提出基于语义的评估方法,缓解BLEU指标的膨胀数字问题,提供更公平评估。
- 在平民术语数据集上训练模型,鼓励模型关注报告的语义内容。
点此查看论文截图