⚠️ 以下所有内容总结都来自于 大语言模型的能力,如有错误,仅供参考,谨慎使用
🔴 请注意:千万不要用于严肃的学术场景,只能用于论文阅读前的初筛!
💗 如果您觉得我们的项目对您有帮助 ChatPaperFree ,还请您给我们一些鼓励!⭐️ HuggingFace免费体验
2025-03-01 更新
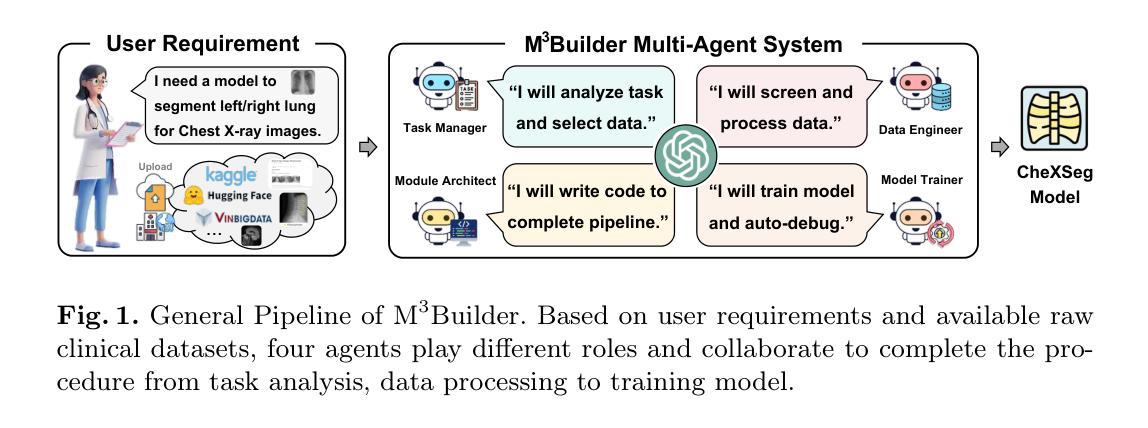
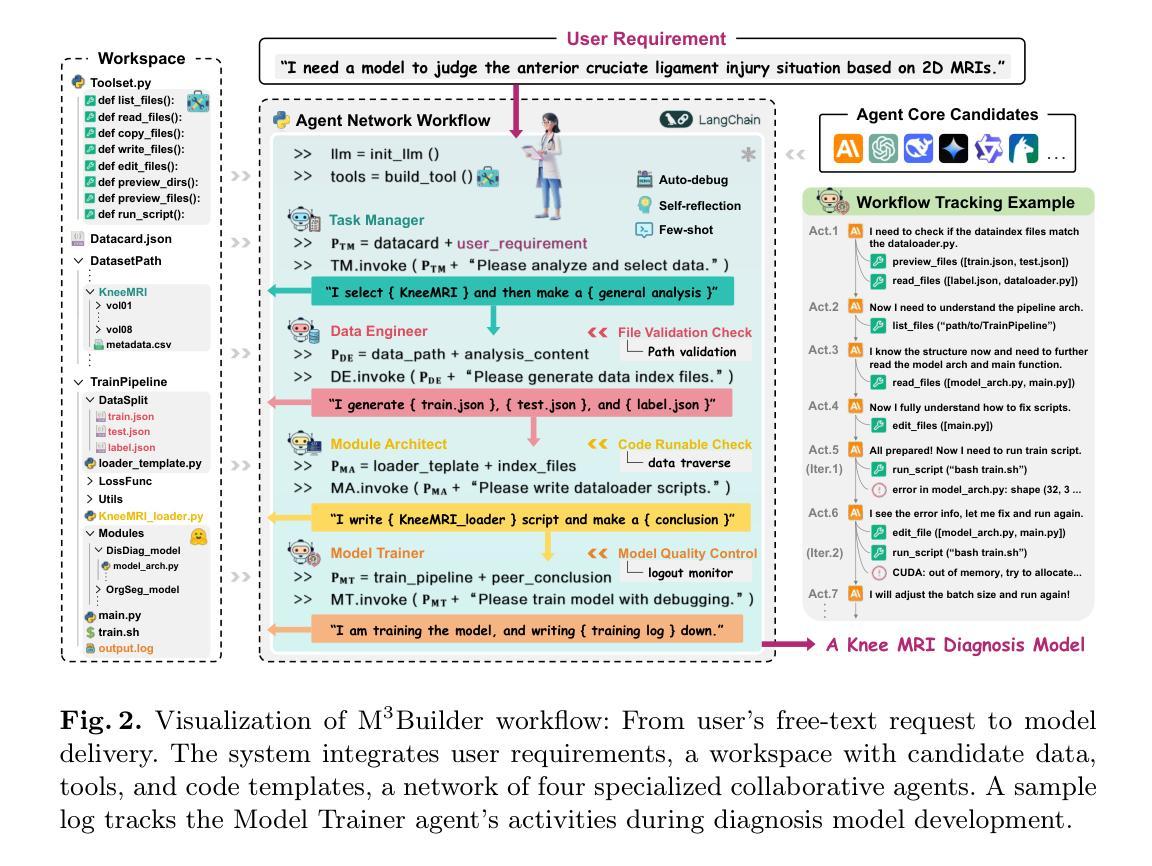
M^3Builder: A Multi-Agent System for Automated Machine Learning in Medical Imaging
Authors:Jinghao Feng, Qiaoyu Zheng, Chaoyi Wu, Ziheng Zhao, Ya Zhang, Yanfeng Wang, Weidi Xie
Agentic AI systems have gained significant attention for their ability to autonomously perform complex tasks. However, their reliance on well-prepared tools limits their applicability in the medical domain, which requires to train specialized models. In this paper, we make three contributions: (i) We present M3Builder, a novel multi-agent system designed to automate machine learning (ML) in medical imaging. At its core, M3Builder employs four specialized agents that collaborate to tackle complex, multi-step medical ML workflows, from automated data processing and environment configuration to self-contained auto debugging and model training. These agents operate within a medical imaging ML workspace, a structured environment designed to provide agents with free-text descriptions of datasets, training codes, and interaction tools, enabling seamless communication and task execution. (ii) To evaluate progress in automated medical imaging ML, we propose M3Bench, a benchmark comprising four general tasks on 14 training datasets, across five anatomies and three imaging modalities, covering both 2D and 3D data. (iii) We experiment with seven state-of-the-art large language models serving as agent cores for our system, such as Claude series, GPT-4o, and DeepSeek-V3. Compared to existing ML agentic designs, M3Builder shows superior performance on completing ML tasks in medical imaging, achieving a 94.29% success rate using Claude-3.7-Sonnet as the agent core, showing huge potential towards fully automated machine learning in medical imaging.
智能体人工智能系统因其自主执行复杂任务的能力而受到广泛关注。然而,它们对精心准备的工具的依赖限制了它们在医学领域的应用,医学领域需要训练专业模型。在本文中,我们做出了三项贡献:(一)我们提出了M3Builder,这是一个新型多智能体系统,旨在自动化医学影像中的机器学习(ML)。M3Builder的核心采用四个专业智能体进行协作,以处理复杂的、多步骤的医学ML工作流程,从自动化数据处理和环境配置到自主的自动调试和模型训练。这些智能体在医学影像ML工作空间内运行,这是一个结构化环境,旨在为智能体提供数据集、训练代码和交互工具的文本描述,从而实现无缝通信和任务执行。(二)为了评估医学影像自动化的ML进度,我们提出了M3Bench,这是一个基准测试,包含四个一般任务在14个训练数据集上,涉及五个解剖部位和三种成像模式,涵盖2D和3D数据。(三)我们尝试了七种最先进的自然语言模型作为我们系统的智能体核心,如Claude系列、GPT-4o和DeepSeek-V3。与现有的ML智能体设计相比,M3Builder在完成医学影像中的ML任务时表现出卓越的性能,使用Claude-3.7-Sonnet作为智能体核心时,成功率达到94.29%,显示出在医学影像全自动机器学习的巨大潜力。
论文及项目相关链接
PDF 38 pages, 7 figures
Summary
本文介绍了M3Builder,一个针对医学成像领域自动化机器学习(ML)的多智能体系统。该系统由四个专门智能体组成,可协作完成复杂的医学ML工作流程,包括数据自动化处理、环境配置、自动调试和模型训练等。此外,本文还提出了M3Bench,一个涵盖不同解剖部位和成像模式的医学成像ML基准测试。实验表明,M3Builder在完成医学成像ML任务方面表现出卓越性能,使用Claude-3.7-Sonnet作为核心智能体的成功率为94.29%。
Key Takeaways
- M3Builder是一个用于医学成像自动化机器学习的多智能体系统,包含四个专门智能体,可协作完成复杂流程。
- M3Builder在医学成像ML任务中表现出卓越性能,使用Claude-3.7-Sonnet作为核心智能体的成功率为94.29%。
- M3Builder在一个结构化的医学成像ML工作空间中运行,提供数据集描述、训练代码和交互工具。
- M3Bench是一个涵盖多种任务和数据集的医学成像ML基准测试,用于评估自动化医学成像ML的进展。
- M3Builder的智能体核心可以采用最新的大型语言模型,如Claude系列、GPT-4o和DeepSeek-V3。
- M3Builder对完全自动化机器学习在医学成像中的潜力具有巨大影响。
点此查看论文截图

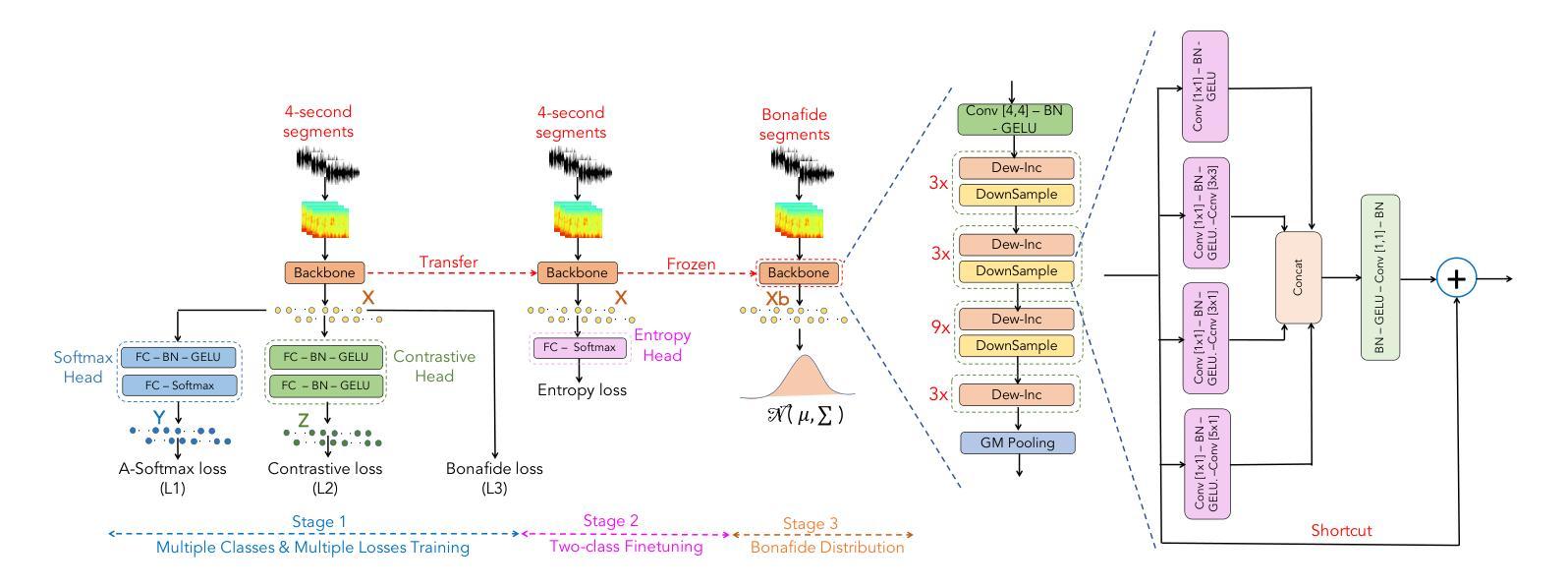
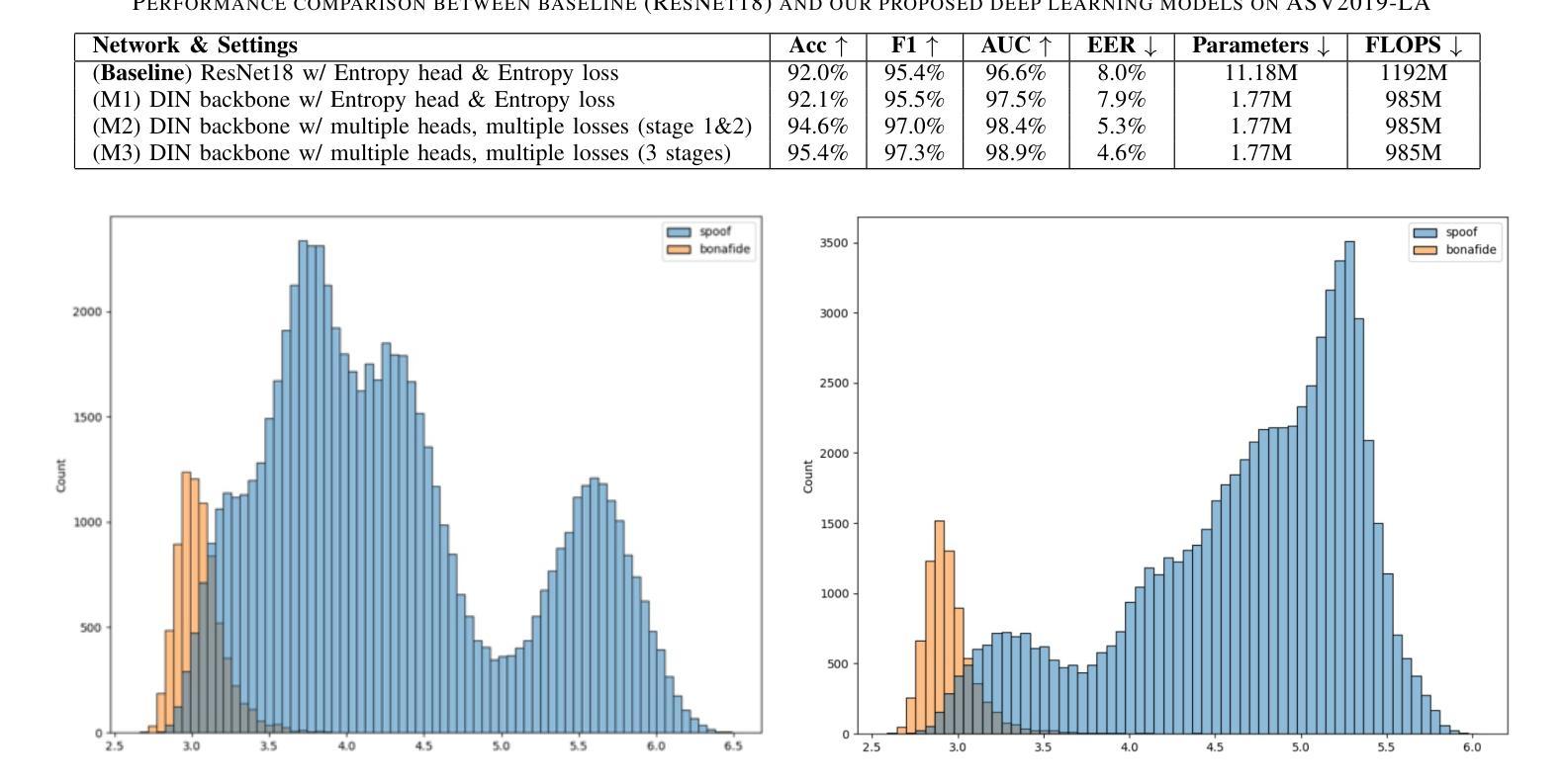
DIN-CTS: Low-Complexity Depthwise-Inception Neural Network with Contrastive Training Strategy for Deepfake Speech Detection
Authors:Lam Pham, Dat Tran, Florian Skopik, Alexander Schindler, Silvia Poletti, Fischinger David, Martin Boyer
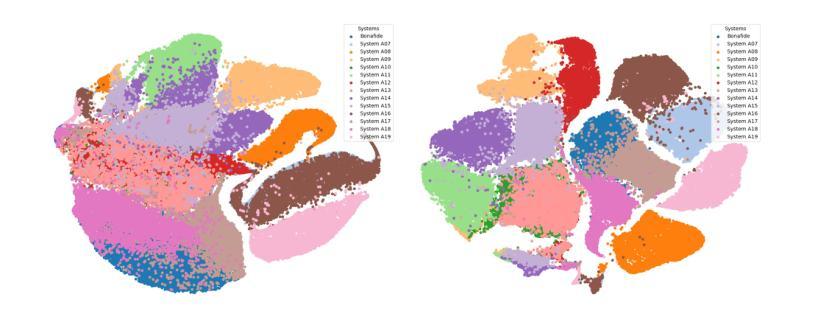
In this paper, we propose a deep neural network approach for deepfake speech detection (DSD) based on a lowcomplexity Depthwise-Inception Network (DIN) trained with a contrastive training strategy (CTS). In this framework, input audio recordings are first transformed into spectrograms using Short-Time Fourier Transform (STFT) and Linear Filter (LF), which are then used to train the DIN. Once trained, the DIN processes bonafide utterances to extract audio embeddings, which are used to construct a Gaussian distribution representing genuine speech. Deepfake detection is then performed by computing the distance between a test utterance and this distribution to determine whether the utterance is fake or bonafide. To evaluate our proposed systems, we conducted extensive experiments on the benchmark dataset of ASVspoof 2019 LA. The experimental results demonstrate the effectiveness of combining the Depthwise-Inception Network with the contrastive learning strategy in distinguishing between fake and bonafide utterances. We achieved Equal Error Rate (EER), Accuracy (Acc.), F1, AUC scores of 4.6%, 95.4%, 97.3%, and 98.9% respectively using a single, low-complexity DIN with just 1.77 M parameters and 985 M FLOPS on short audio segments (4 seconds). Furthermore, our proposed system outperforms the single-system submissions in the ASVspoof 2019 LA challenge, showcasing its potential for real-time applications.
本文提出了一种基于低复杂度深度可分离感知网络(DIN)和对比训练策略(CTS)的深度伪造语音检测(DSD)的深度神经网络方法。在该框架中,首先使用短时傅里叶变换(STFT)和线性滤波器(LF)将输入音频记录转换为频谱图,然后用它们来训练DIN。训练完成后,DIN处理真实语音片段以提取音频嵌入,用于构建代表真实语音的高斯分布。然后通过计算测试语音片段与此分布之间的距离来进行深度伪造检测,以确定语音片段是伪造的还是真实的。为了评估我们提出的系统,我们在ASVspoof 2019 LA的基准数据集上进行了大量实验。实验结果证明了深度可分离感知网络与对比学习策略相结合在区分伪造和真实语音片段方面的有效性。使用单一、低复杂度的DIN,在短音频片段(4秒)上,我们实现了等误率(EER)、准确率(Acc.)、F1、AUC得分分别为4.6%、95.4%、97.3%和98.9%。此外,我们提出的系统在ASVspoof 2019 LA挑战中的单系统提交中表现突出,展示了其在实时应用中的潜力。
论文及项目相关链接
Summary
本文提出了一种基于深度神经网络的深度伪造语音检测(DSD)方法,该方法使用低复杂度的Depthwise-Inception网络(DIN)和对比训练策略(CTS)。音频输入首先通过短时傅里叶变换(STFT)和线性滤波器(LF)转换为频谱图来训练DIN。训练后的DIN处理真实语音以提取音频嵌入,构建代表真实语音的高斯分布。通过计算测试语音与此分布之间的距离来进行深度伪造检测,以确定语音是否真实。在ASVspoof 2019 LA基准数据集上进行的实验证明了Depthwise-Inception网络与对比学习策略相结合的有效性。使用单一、低复杂度的DIN,在短短4秒的音频片段上,我们取得了4.6%的等错误率(EER)、95.4%的准确率(Acc.)、97.3%的F1分数和98.9%的AUC分数。此外,我们的系统在ASVspoof 2019 LA挑战中的单系统提交中表现最佳,展示了其实时应用的潜力。
Key Takeaways
- 论文提出了一种基于深度神经网络的深度伪造语音检测方法。
- 使用低复杂度的Depthwise-Inception网络(DIN)和对比训练策略(CTS)。
- 输入音频通过短时傅里叶变换和线性滤波器转换为频谱图进行训练。
- DIN能够处理真实语音并提取音频嵌入,构建代表真实语音的高斯分布。
- 通过计算测试语音与高斯分布之间的距离来判断其真伪。
- 在ASVspoof 2019 LA数据集上的实验证明了该方法的有效性。
点此查看论文截图


Generative augmentations for improved cardiac ultrasound segmentation using diffusion models
Authors:Gilles Van De Vyver, Aksel Try Lenz, Erik Smistad, Sindre Hellum Olaisen, Bjørnar Grenne, Espen Holte, Håavard Dalen, Lasse Løvstakken
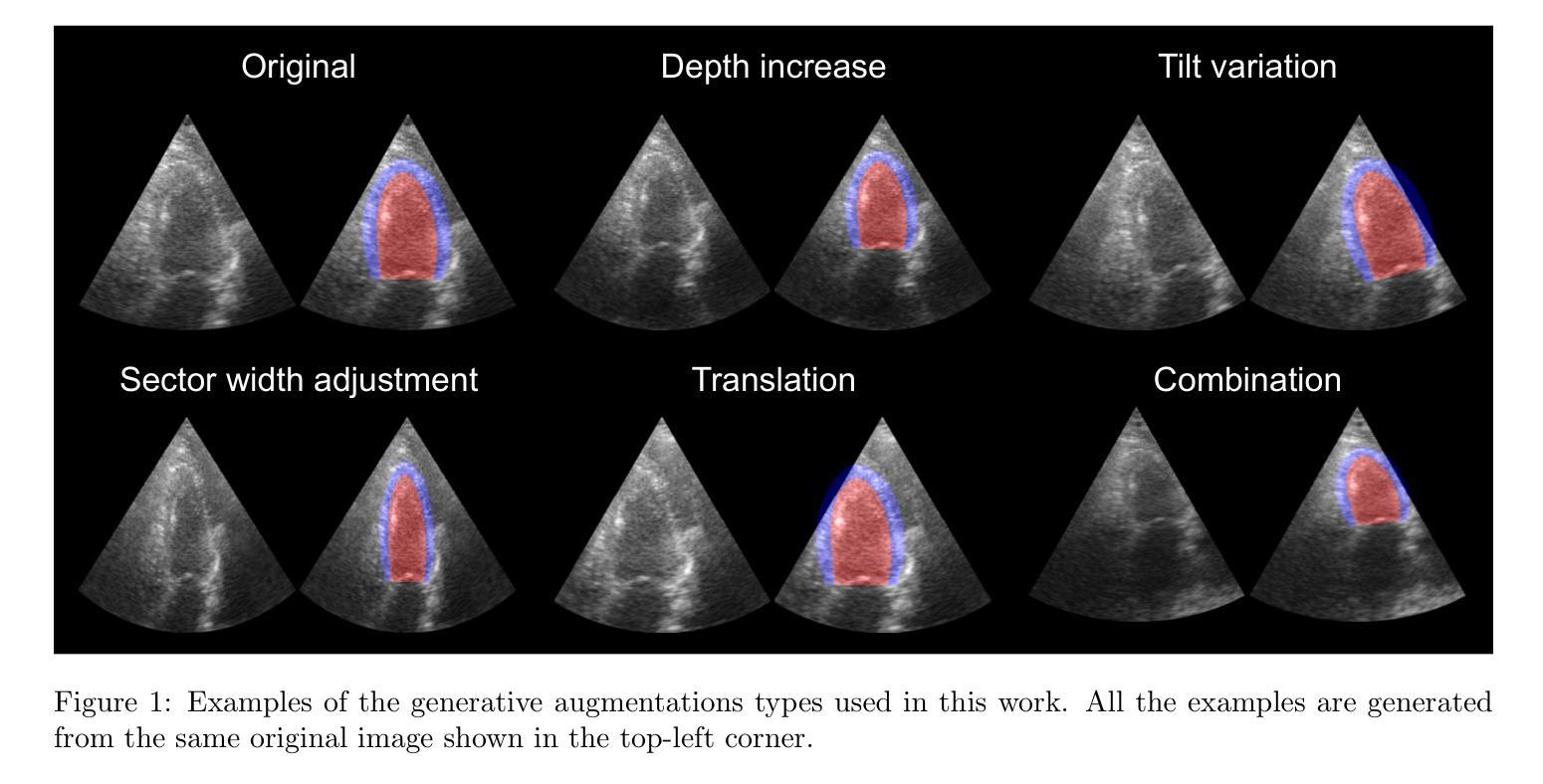
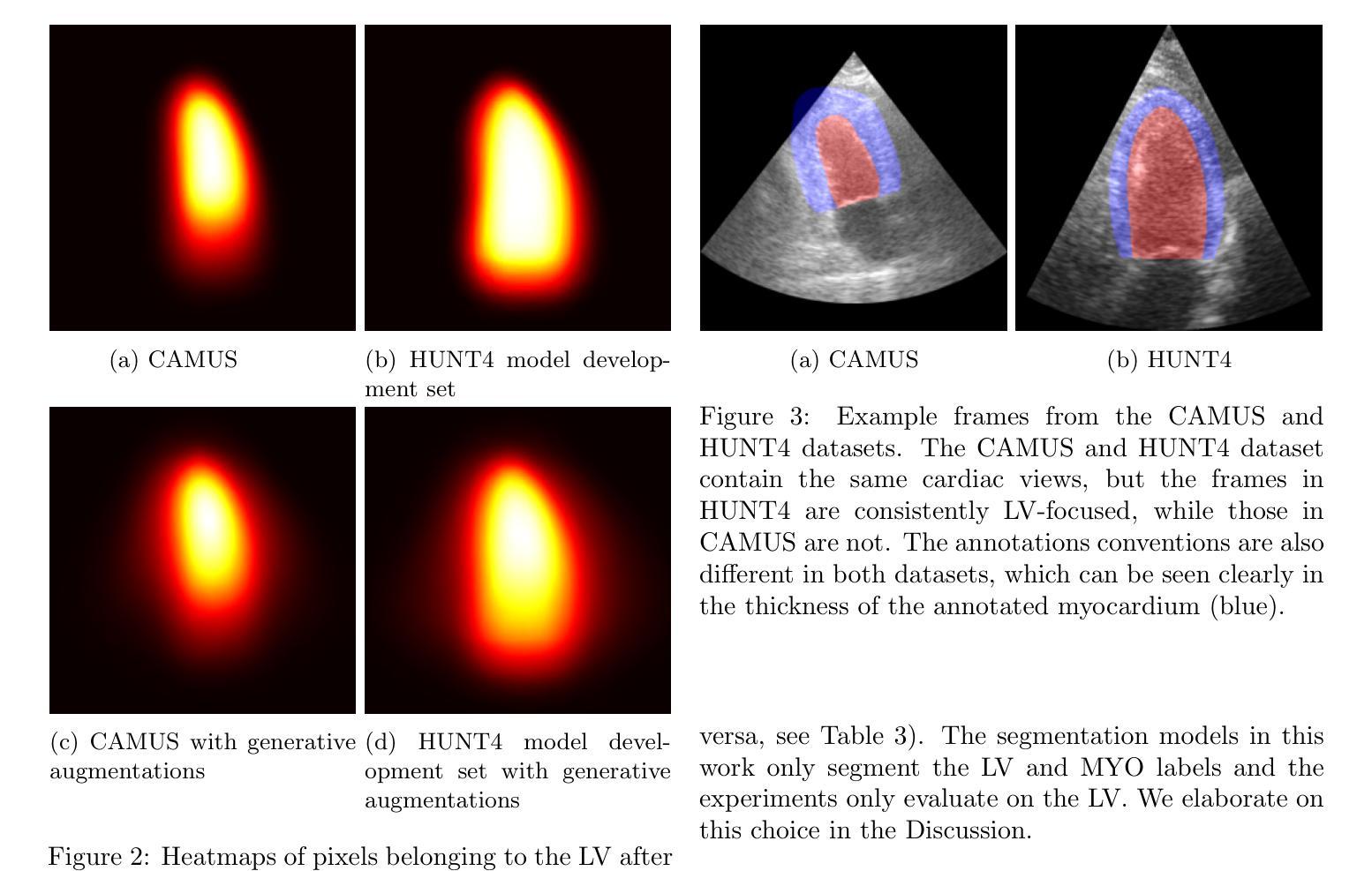
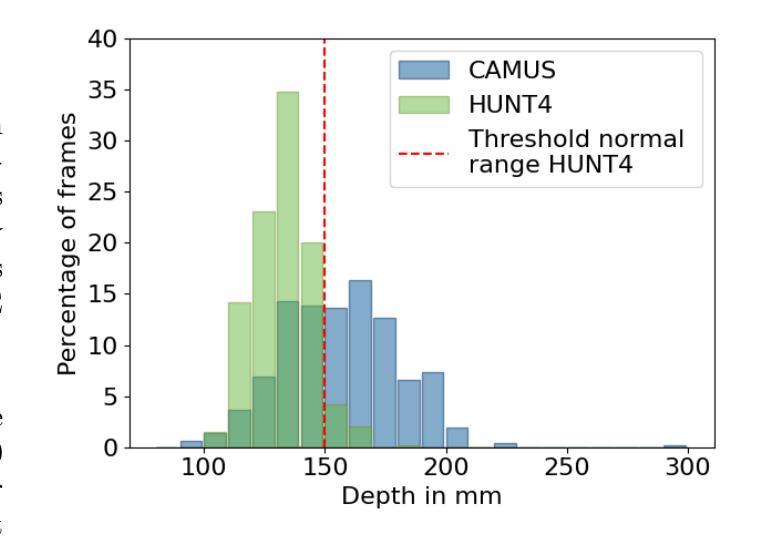
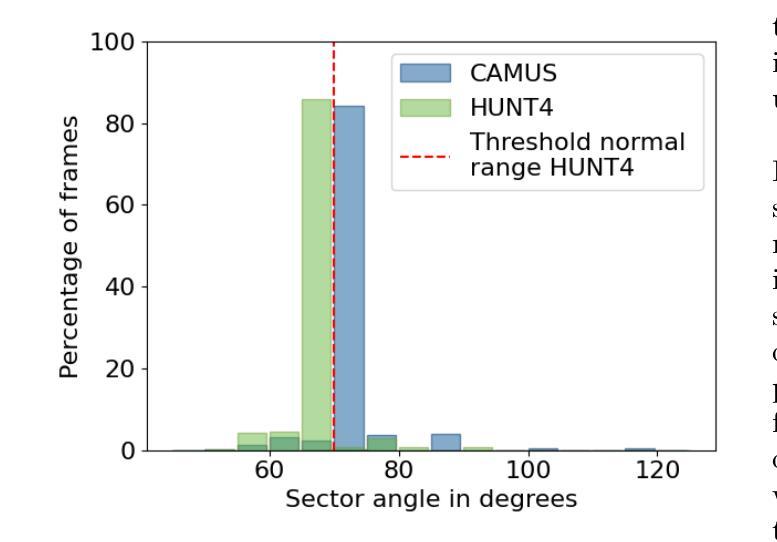
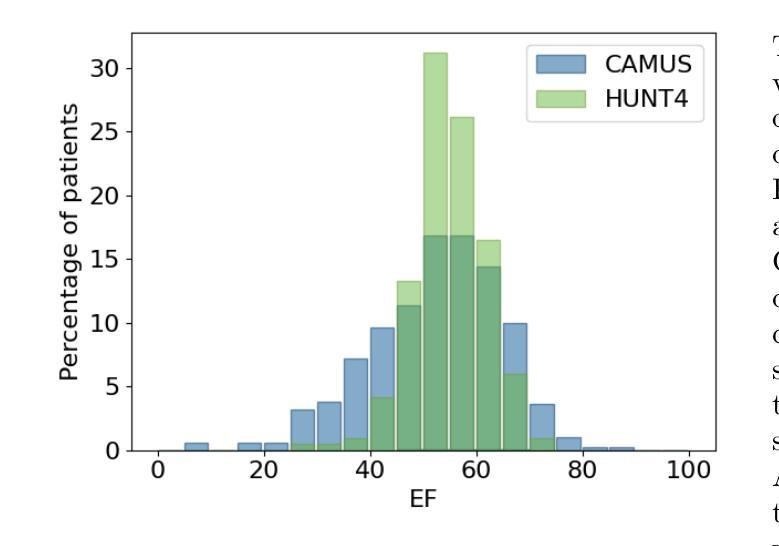
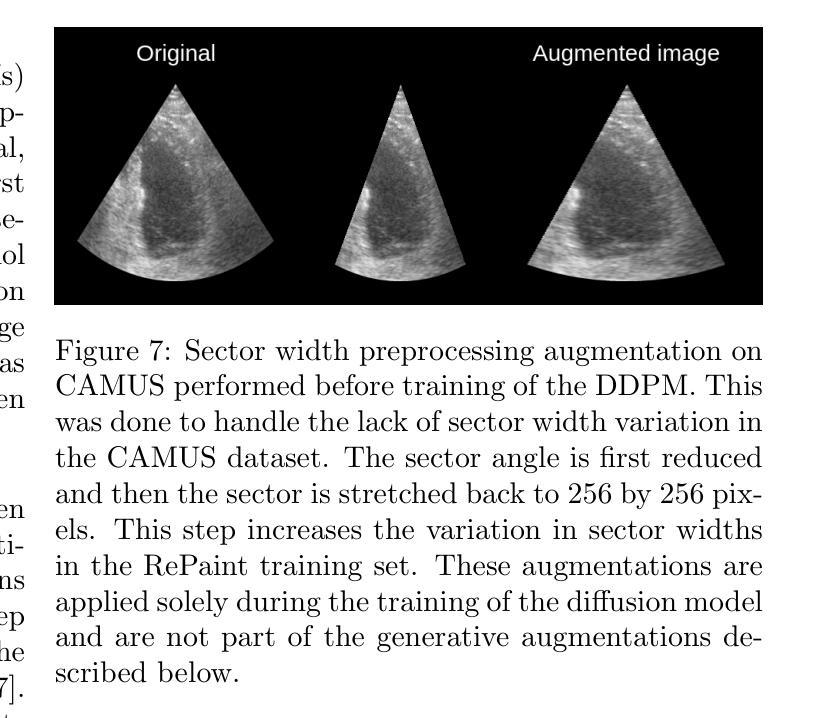
One of the main challenges in current research on segmentation in cardiac ultrasound is the lack of large and varied labeled datasets and the differences in annotation conventions between datasets. This makes it difficult to design robust segmentation models that generalize well to external datasets. This work utilizes diffusion models to create generative augmentations that can significantly improve diversity of the dataset and thus the generalisability of segmentation models without the need for more annotated data. The augmentations are applied in addition to regular augmentations. A visual test survey showed that experts cannot clearly distinguish between real and fully generated images. Using the proposed generative augmentations, segmentation robustness was increased when training on an internal dataset and testing on an external dataset with an improvement of over 20 millimeters in Hausdorff distance. Additionally, the limits of agreement for automatic ejection fraction estimation improved by up to 20% of absolute ejection fraction value on out of distribution cases. These improvements come exclusively from the increased variation of the training data using the generative augmentations, without modifying the underlying machine learning model. The augmentation tool is available as an open source Python library at https://github.com/GillesVanDeVyver/EchoGAINS.
当前心脏超声分割研究面临的主要挑战之一是缺乏大规模且多样化的标记数据集以及不同数据集之间标注规范的不同。这使得设计能够良好地推广到外部数据集的稳健分割模型变得困难。本研究利用扩散模型创建生成增强技术,可以显著提高数据集的多样性,从而在无需更多注释数据的情况下提高分割模型的可推广性。除了常规增强技术外,还应用了这些生成增强技术。视觉测试调查表明,专家无法清楚区分真实图像和完全生成的图像。使用所提出的生成增强技术,在内部数据集上进行训练并在外部数据集上进行测试时,分割稳健性有所提高,豪斯多夫距离提高了超过20毫米。此外,在超出分布范围的案例中,自动射血分数估计的协议限制提高了高达绝对射血分数值的20%。这些改进完全来自于使用生成增强技术后训练数据变异的增加,而没有修改基础机器学习模型。该增强工具可作为开源Python库在https://github.com/GillesVanDeVyver/EchoGAINS获取。
论文及项目相关链接
Summary
本文利用扩散模型创建生成性增强数据,有效提高了数据集多样性,从而提高了分割模型的泛化能力,即使在没有更多注释数据的情况下也能实现。通过应用常规增强和生成性增强,专家无法明确区分真实和完全生成的图像。在内部数据集上进行训练、在外部数据集上进行测试时,提高了分割的稳健性,豪斯洛距离提高了20毫米以上。此外,在自动射血分数估计方面,对超出分布范围的案例,绝对射血分数值的同意限度提高了20%。这些改进完全来自于使用生成增强数据的训练数据多样性的增加,而没有修改基础机器学习模型。
Key Takeaways
- 当前心脏超声分割研究的主要挑战之一是缺乏大规模、多样化的标记数据集以及数据集之间注释规范的不同。
- 扩散模型被用来创建生成性增强数据,以提高数据集的多样性和分割模型的泛化能力。
- 生成性增强数据的应用使得专家难以区分真实和生成的图像。
- 在内部数据集上训练、在外部数据集上测试时,使用生成性增强数据提高了分割稳健性,表现在豪斯洛距离的提高上。
- 自动射血分数估计的改进显著,对超出分布范围的案例,绝对射血分数值的同意限度提高了20%。
- 这些改进来源于训练数据多样性的增加,而不是对基础机器学习模型的修改。
点此查看论文截图


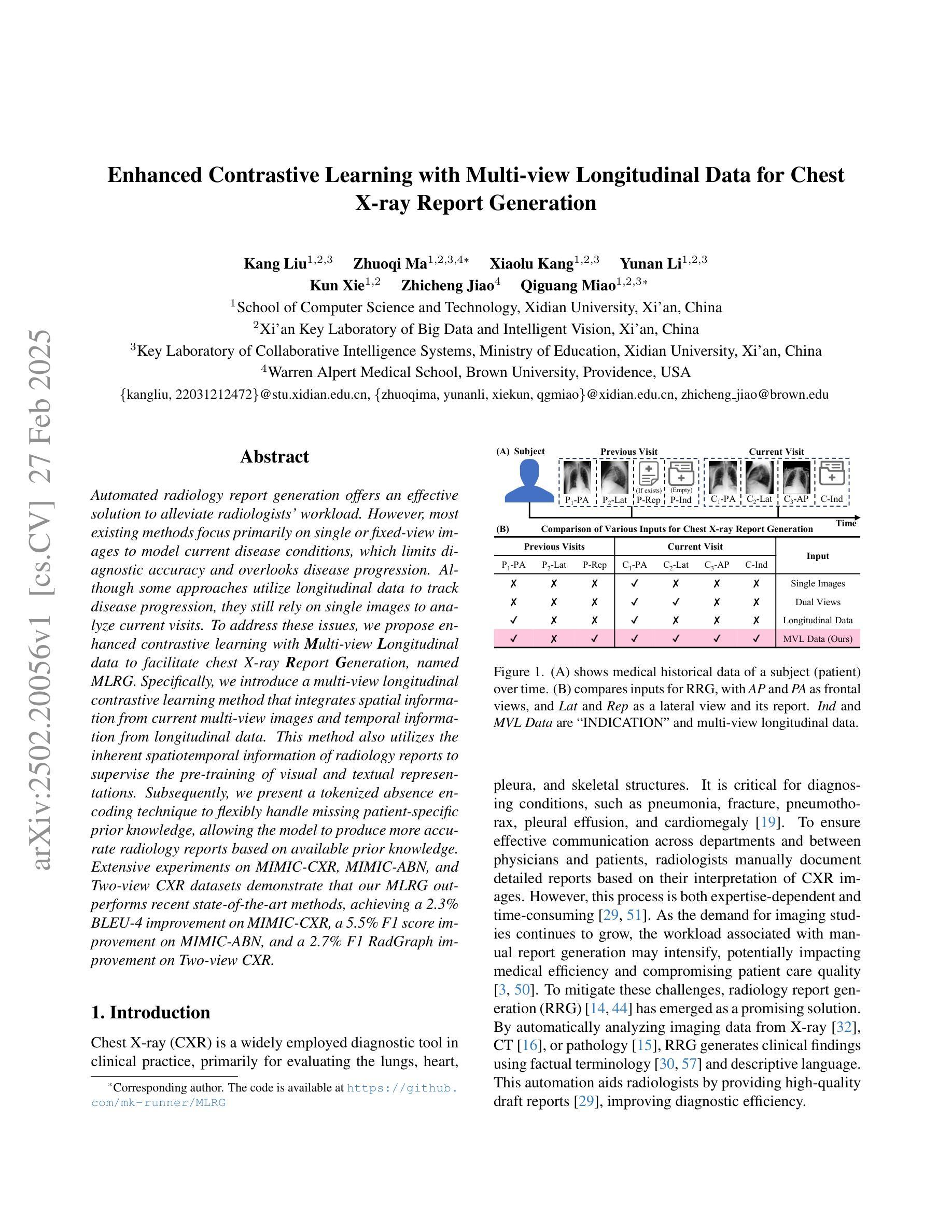
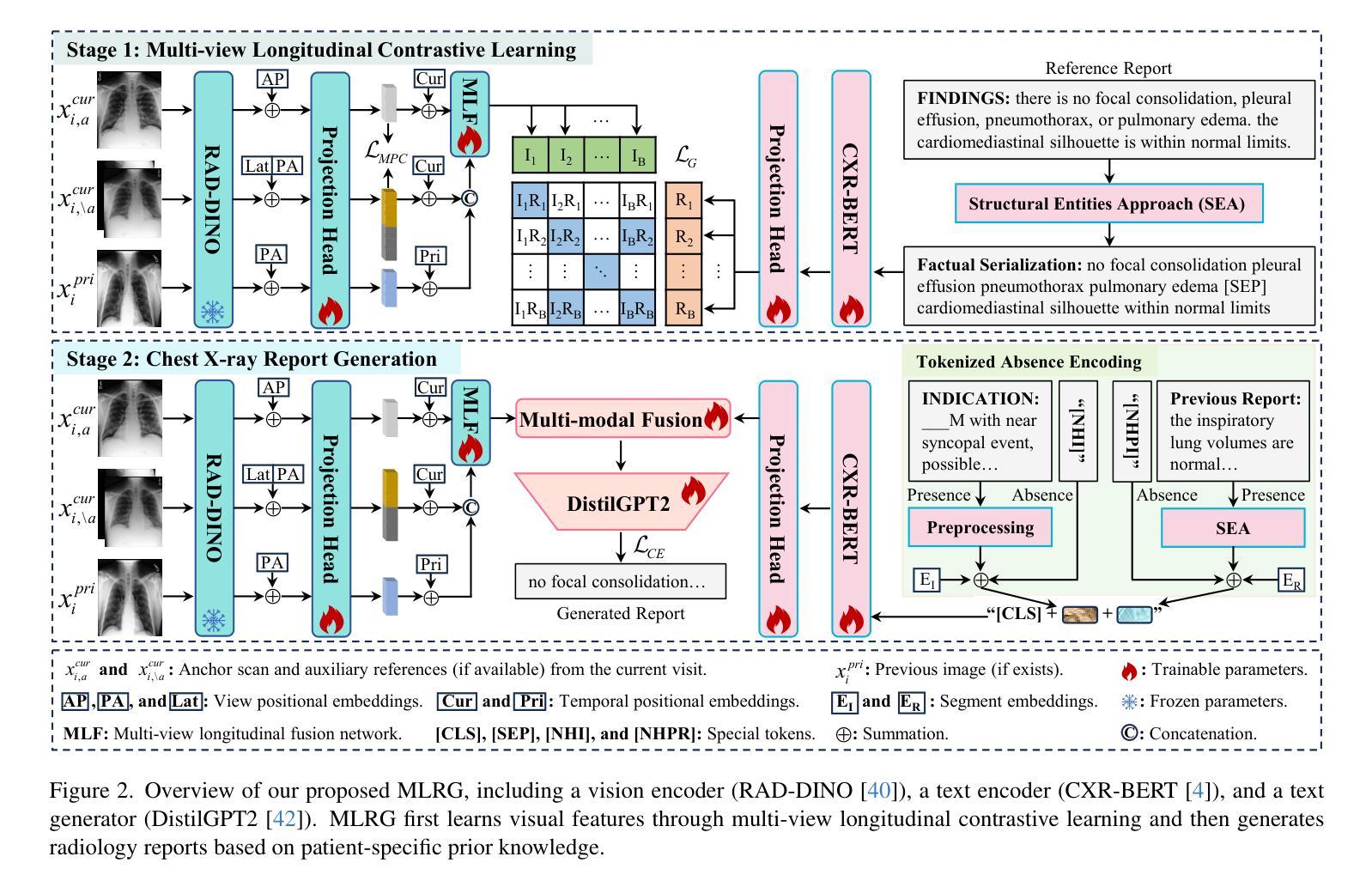
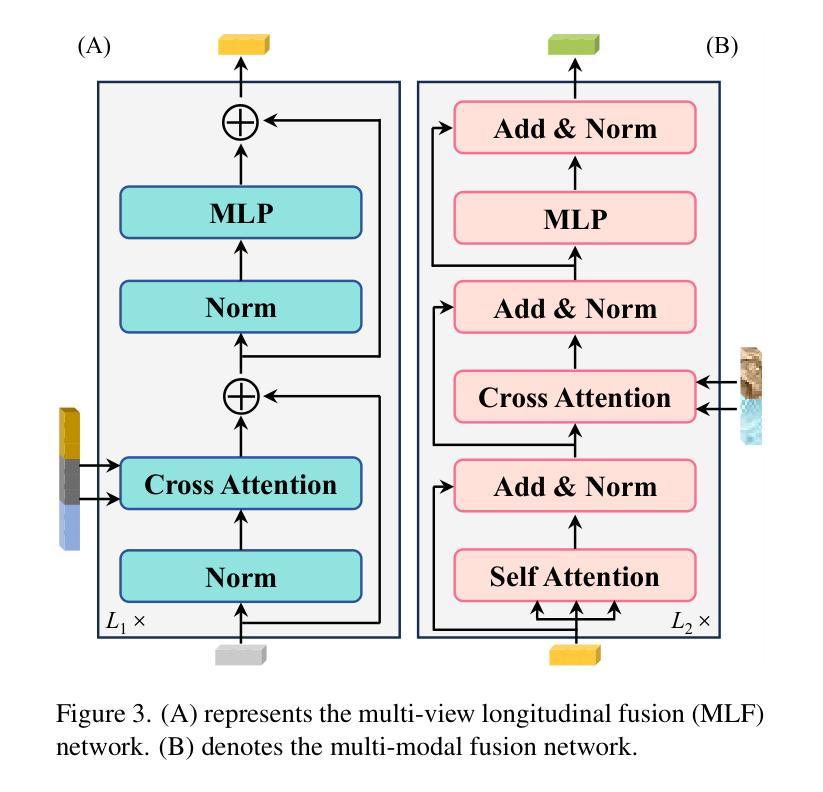
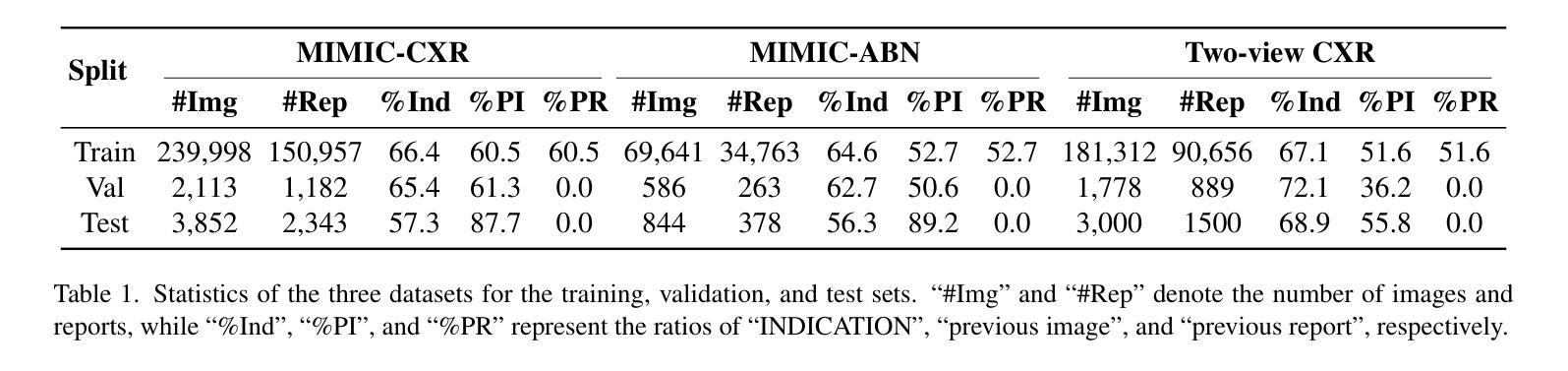
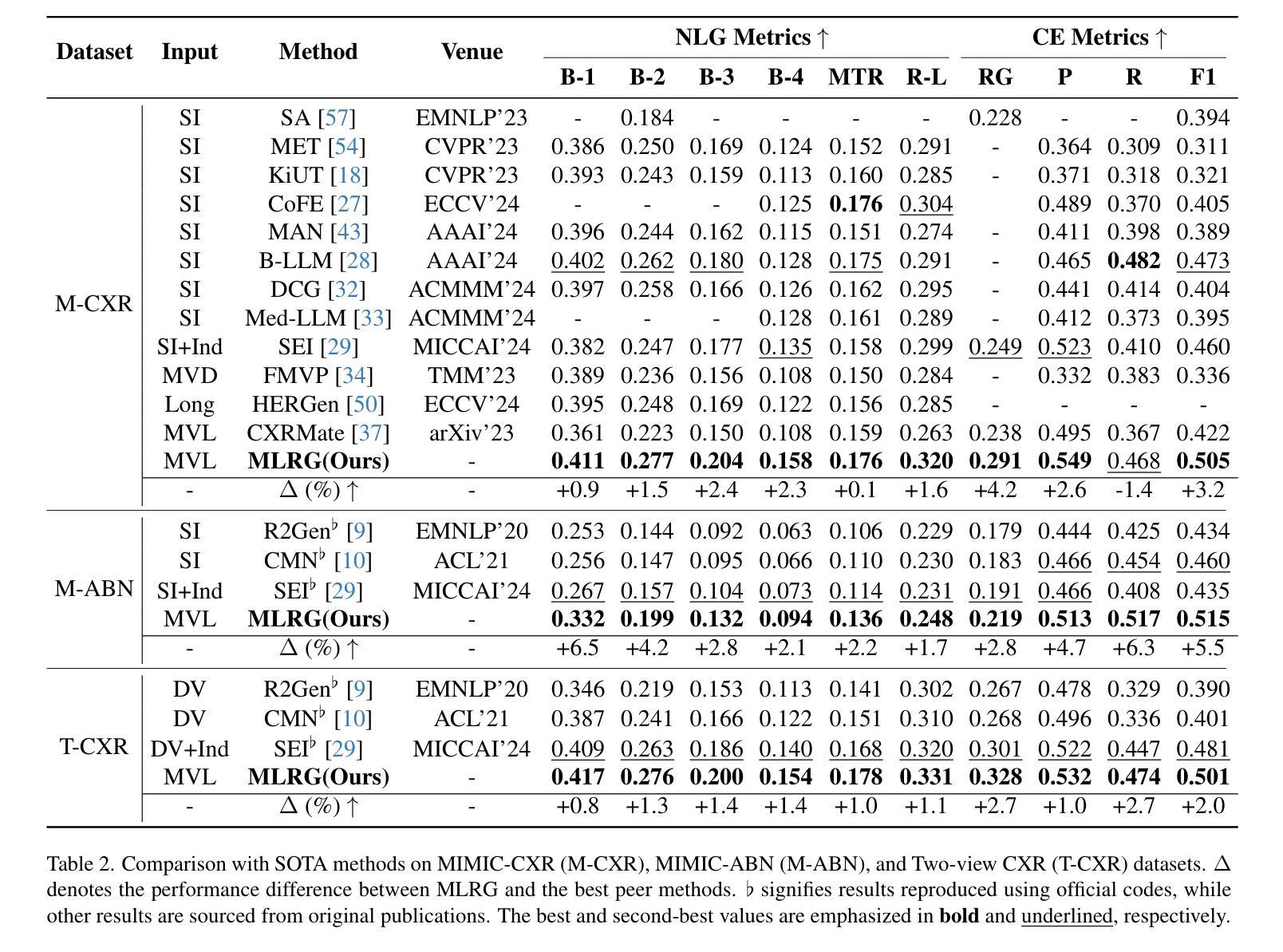
Enhanced Contrastive Learning with Multi-view Longitudinal Data for Chest X-ray Report Generation
Authors:Kang Liu, Zhuoqi Ma, Xiaolu Kang, Yunan Li, Kun Xie, Zhicheng Jiao, Qiguang Miao
Automated radiology report generation offers an effective solution to alleviate radiologists’ workload. However, most existing methods focus primarily on single or fixed-view images to model current disease conditions, which limits diagnostic accuracy and overlooks disease progression. Although some approaches utilize longitudinal data to track disease progression, they still rely on single images to analyze current visits. To address these issues, we propose enhanced contrastive learning with Multi-view Longitudinal data to facilitate chest X-ray Report Generation, named MLRG. Specifically, we introduce a multi-view longitudinal contrastive learning method that integrates spatial information from current multi-view images and temporal information from longitudinal data. This method also utilizes the inherent spatiotemporal information of radiology reports to supervise the pre-training of visual and textual representations. Subsequently, we present a tokenized absence encoding technique to flexibly handle missing patient-specific prior knowledge, allowing the model to produce more accurate radiology reports based on available prior knowledge. Extensive experiments on MIMIC-CXR, MIMIC-ABN, and Two-view CXR datasets demonstrate that our MLRG outperforms recent state-of-the-art methods, achieving a 2.3% BLEU-4 improvement on MIMIC-CXR, a 5.5% F1 score improvement on MIMIC-ABN, and a 2.7% F1 RadGraph improvement on Two-view CXR.
自动放射学报告生成提供了一种有效的解决方案,以减轻放射科医师的工作量。然而,大多数现有方法主要关注单视图或固定视图图像来建模当前疾病状况,这限制了诊断的准确性并忽略了疾病的进展。虽然有些方法利用纵向数据追踪疾病进展,但它们仍然依赖于单幅图像来分析当前就诊情况。为了解决这些问题,我们提出了使用多视图纵向数据的增强对比学习来促进胸部X射线报告生成,命名为MLRG。具体来说,我们引入了一种多视图纵向对比学习方法,该方法结合了当前多视图图像的空间信息和纵向数据的时序信息。这种方法还利用放射学报告中的固有时空信息来监督视觉和文本表示的预训练。随后,我们提出了一种标记缺失编码技术,可以灵活地处理特定于患者的缺失先验知识,使模型能够根据可用的先验知识生成更准确的放射学报告。在MIMIC-CXR、MIMIC-ABN和Two-view CXR数据集上的大量实验表明,我们的MLRG优于最新的先进技术,在MIMIC-CXR上BLEU-4提高了2.3%,在MIMIC-ABN上F1得分提高了5.5%,在Two-view CXR上F1 RadGraph提高了2.7%。
论文及项目相关链接
PDF Accepted by CVPR 2025
Summary
基于医学图像自动化生成报告能有效缓解放射科医生的工作负担,但现有方法主要关注单一或固定视角的图像来建模当前疾病状况,限制了诊断准确性和疾病进展的洞察。本研究提出了基于多视角纵向数据的对比学习增强方法,用于促进胸部X光报告生成(命名为MLRG)。该方法结合了当前多视角图像的空间信息和纵向数据的时序信息,并利用放射学报告中的时空信息对视觉和文本表示进行预训练。此外,还提出了一种标记缺失编码技术,可灵活处理患者特定先验知识的缺失,使模型基于可用的先验知识产生更准确的放射学报告。实验表明,MLRG在MIMIC-CXR、MIMIC-ABN和Two-view CXR数据集上的表现优于最新先进技术,在MIMIC-CXR上BLEU-4提高了2.3%,在MIMIC-ABN上F1分数提高了5.5%,在Two-view CXR上F1 RadGraph提高了2.7%。
Key Takeaways
- 自动化生成医学图像报告可以缓解放射科医生的工作负担。
- 当前方法在诊断准确性和疾病进展洞察方面存在局限性,主要关注单一或固定视角的图像。
- 提出了一种基于多视角纵向数据的对比学习方法MLRG,结合空间和时序信息。
- 利用放射学报告的时空信息对视觉和文本表示进行预训练。
- 提出了标记缺失编码技术,以灵活处理患者特定先验知识的缺失。
- MLRG在多个数据集上的表现优于现有技术。
点此查看论文截图
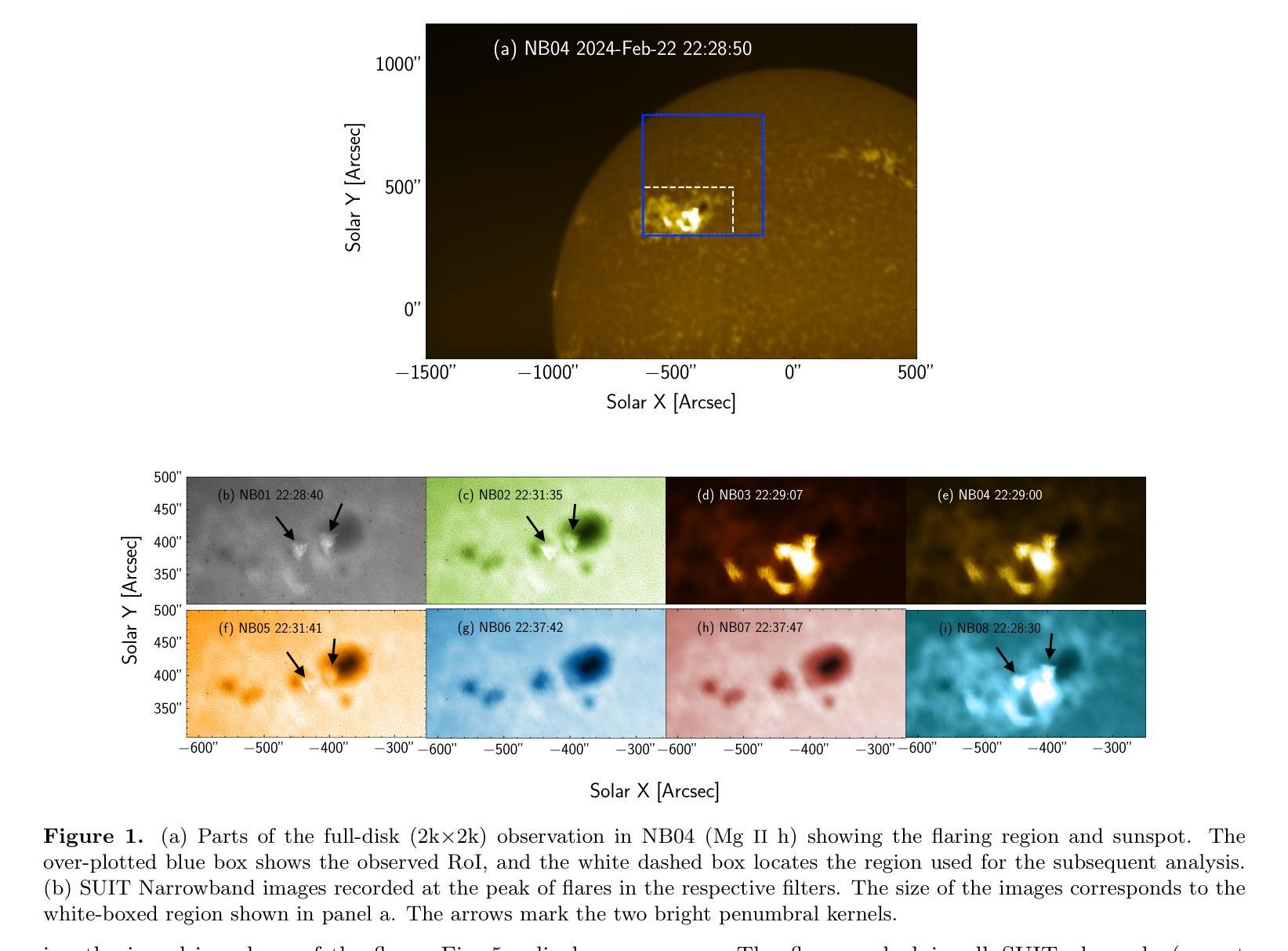
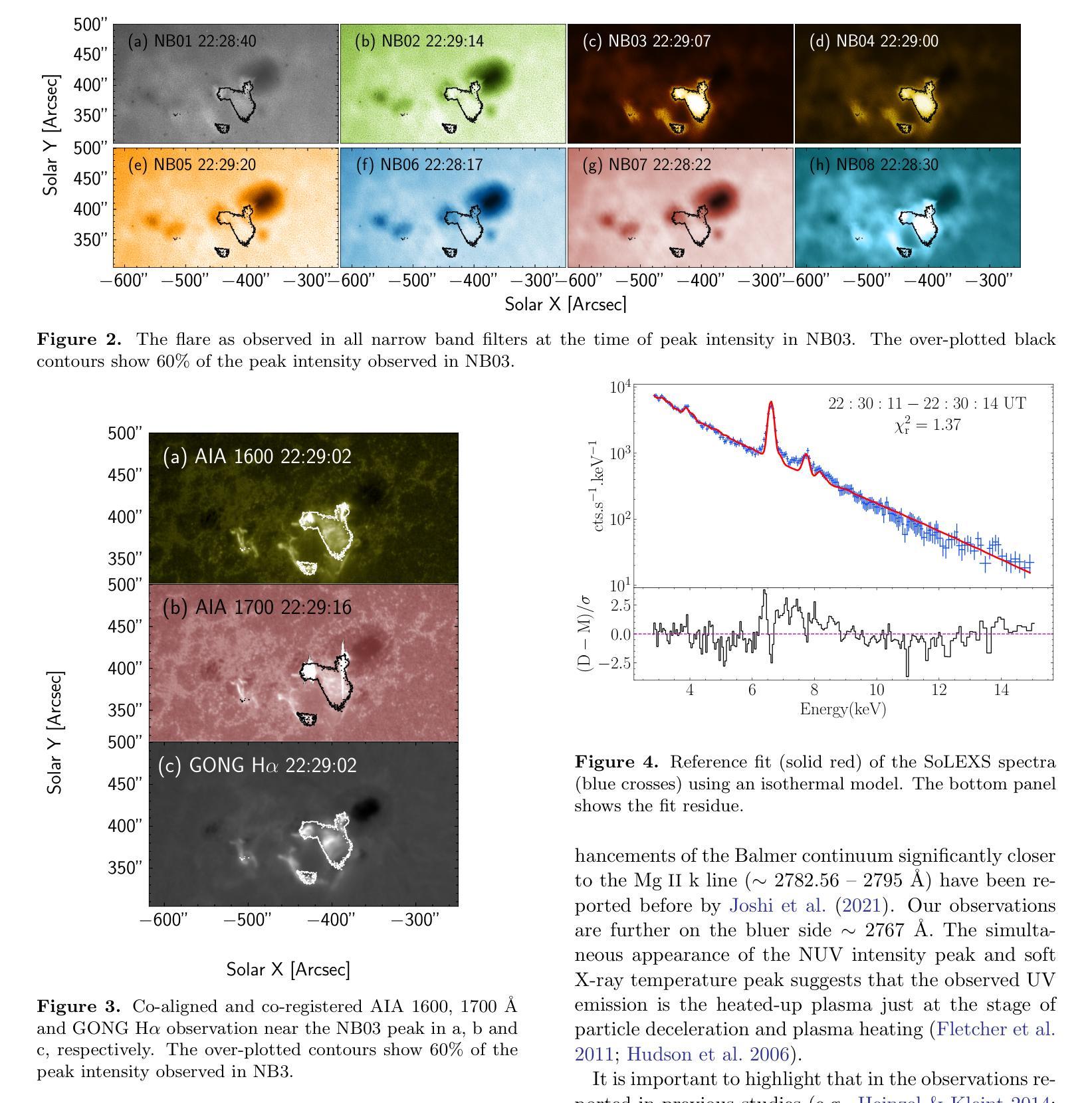
Near and Mid UltraViolet Observations of X-6.3 flare on 22nd February 2024 recorded by the Solar Ultraviolet Imaging Telescope on board Aditya-L1
Authors:Soumya Roy, Durgesh Tripathi, Sreejith Padinhatteeri, A. N. Ramaprakash, Abhilash R. Sarwade, V. N. Nived, Janmejoy Sarkar, Rahul Gopalakrishnan, Rushikesh Deogaonkar, K. Sankarasubramanian, Sami K. Solanki, Dibyendu Nandy, Dipankar Banerjee
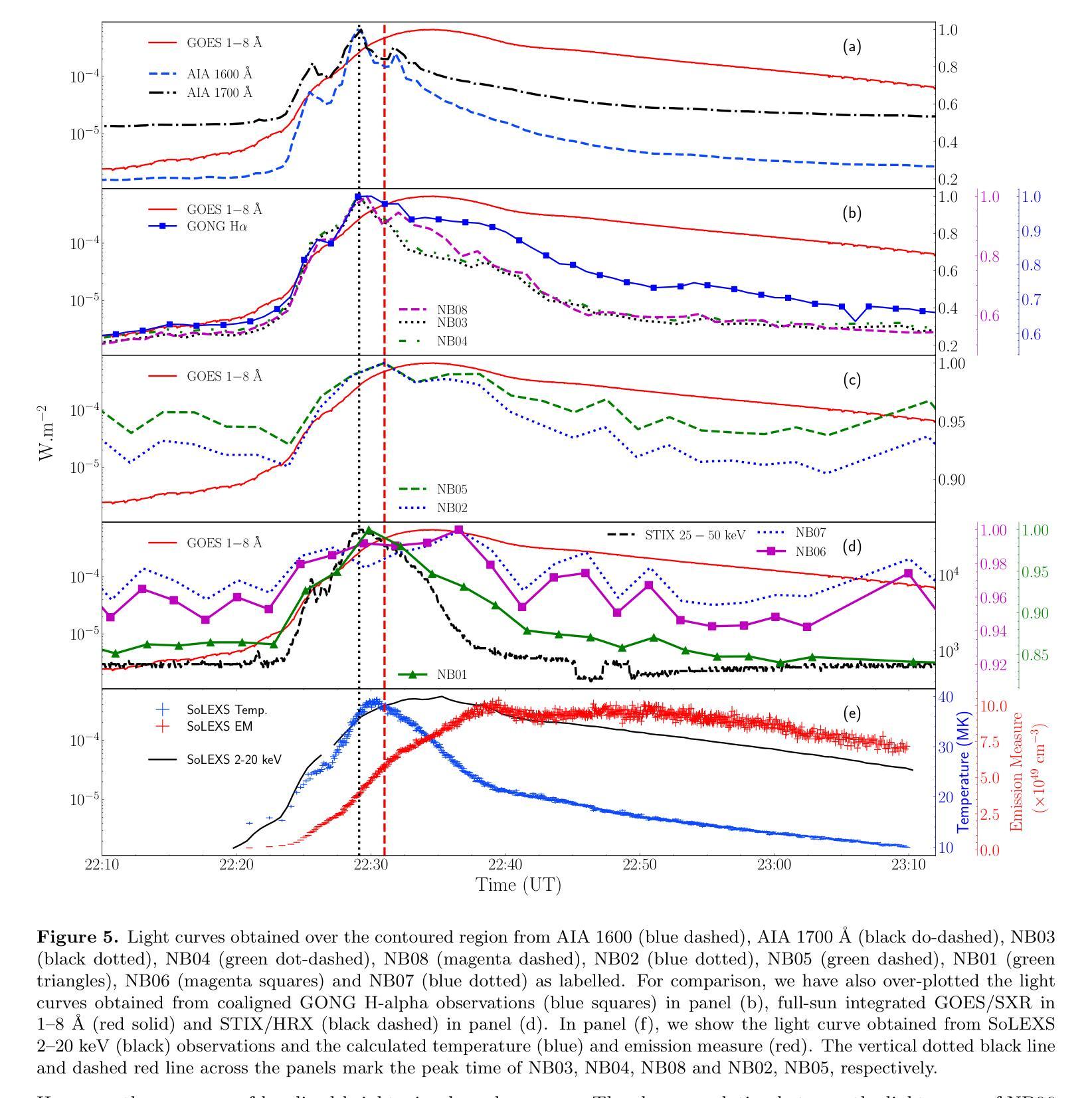
Solar flares are regularly observed in extreme ultraviolet (EUV), soft X-rays (SXR), and hard X-rays (HXR). However, those in near and mid-UV are sparse. The Solar Ultraviolet Imaging Telescope (SUIT) onboard the Aditya-L1, launched on 2nd September, 2023 provides regular observations in the 200-400 nm wavelength range through eleven filters. Here, we report the observation of the X6.3 flare on Feb 22, 2024 using eight narrow band (NB) filters of SUIT. We have also used co-spatiotemporal observations from SDO/AIA, Solar Orbiter/STIX, GONG H$\alpha$, Aditya-L1/SoLEXS and GOES. We obtained light curves over the flaring region from AIA 1600, 1700 \r{A} and GONG H$\alpha$ and compared them with the disk-integrated lightcurve obtained from GOES and SoLEXS SXR and STIX HXR. We find that the flare peaks in SUIT NB01, NB03, NB04, and NB08 filters simultaneously with HXR, 1600, and 1700 \r{A} along with the peak temperature obtained from SoLEXS. In contrast, in NB02 and NB05, the flare peaks $\sim$ 2 minutes later than the HXR peak, while in NB06 and NB07, the flare peaks $\sim$ 3 minutes after the GOES soft X-ray peak. To the best of our knowledge, this is the first observation of a flare in these wavelengths (except in NB03, NB04 and NB05). Moreover, for the first time, we show the presence of a bright kernel in NB02. These results demonstrate the capabilities of SUIT observations in flare studies.
太阳耀斑经常会在极紫外(EUV)、软X射线(SXR)和硬X射线(HXR)中观察到。然而,在近紫外和中紫外中的耀斑则较为稀少。Aditya-L1号卫星上的太阳能紫外线成像望远镜(SUIT)于2023年9月2日发射升空,可以通过十一个滤镜提供200-400纳米波长范围内的定期观测。在这里,我们报告了使用SUIT的八个窄带(NB)滤镜观察到的2024年2月22日的X6.3级耀斑。我们还使用了来自SDO/AIA、Solar Orbiter/STIX、GONG Hα、Aditya-L1/SoLEXS和GOES的协同时空观测数据。我们从AIA 1600、1700Å和GONG Hα获得了耀斑区的光变曲线,并与来自GOES和SoLEXS SXR以及STIX HXR的盘集成光变曲线进行了比较。我们发现,在SUIT的NB01、NB03、NB04和NB08滤镜中,耀斑峰值与HXR、1600和1700Å同时出现,并伴随着由SoLEXS获得的峰值温度。相比之下,在NB02和NB05中,耀斑峰值比HXR峰值晚了约2分钟,而在NB06和NB07中,耀斑峰值则比GOES软X射线峰值晚了约3分钟。据我们所知,这是(除NB03、NB04和NB05外)首次在这些波长下观察到的耀斑。而且,我们首次在NB02中发现了明亮的内核。这些结果展示了SUIT在耀斑研究中的观测能力。
论文及项目相关链接
PDF Accepted for publication in ApJL, 9 pages, 5 figures
Summary
太阳耀斑在极紫外波段(EUV)、软X射线(SXR)和硬X射线(HXR)中经常观测到,但在近紫外和中紫外观测稀少。利用太阳紫外成像望远镜(SUIT)的观察,可在特定的八个窄带滤波器中对特定耀斑进行检测分析。与其他设备的协同观察结果表明,耀斑的特性在特定的滤光器中与特定光波段之间存在某种一致性或特定关联。这是首次在这些波长(除NB03、NB04和NB05外)观测到的耀斑,展示了SUIT观测在耀斑研究中的潜力。
Key Takeaways
- 太阳耀斑在不同波长下的观测结果有所不同,尤其是近紫外和中紫外波段相对较少被观察到。
- Aditya-L1上的Solar Ultraviolet Imaging Telescope(SUIT)能够在特定的八个窄带滤波器中对太阳耀斑进行详细的观察分析。
- 在特定滤光器下的耀斑峰值与特定光波段之间存在同步现象。例如,某些滤镜捕捉到的耀斑峰值与硬X射线峰值同步出现。
- 在某些滤镜中,如NB02和NB05,太阳耀斑峰值比硬X射线峰值延迟约2分钟或更长的时间出现。这种差异表明太阳耀斑在不同波长下的动态行为具有复杂性。
- SUIT观测结果揭示了首次观察到的特定波长下的太阳耀斑特性,尤其是在某些窄带滤波器中。这显示了SUIT在太阳耀斑研究中的独特价值。
- 本次观测还首次发现了名为“亮核”的现象存在于特定的滤光器中,如NB02。这为太阳耀斑的进一步研究提供了新的视角和研究方向。
点此查看论文截图

Deep Learning-Based Approach for Automatic 2D and 3D MRI Segmentation of Gliomas
Authors:Kiranmayee Janardhan, Christy Bobby T
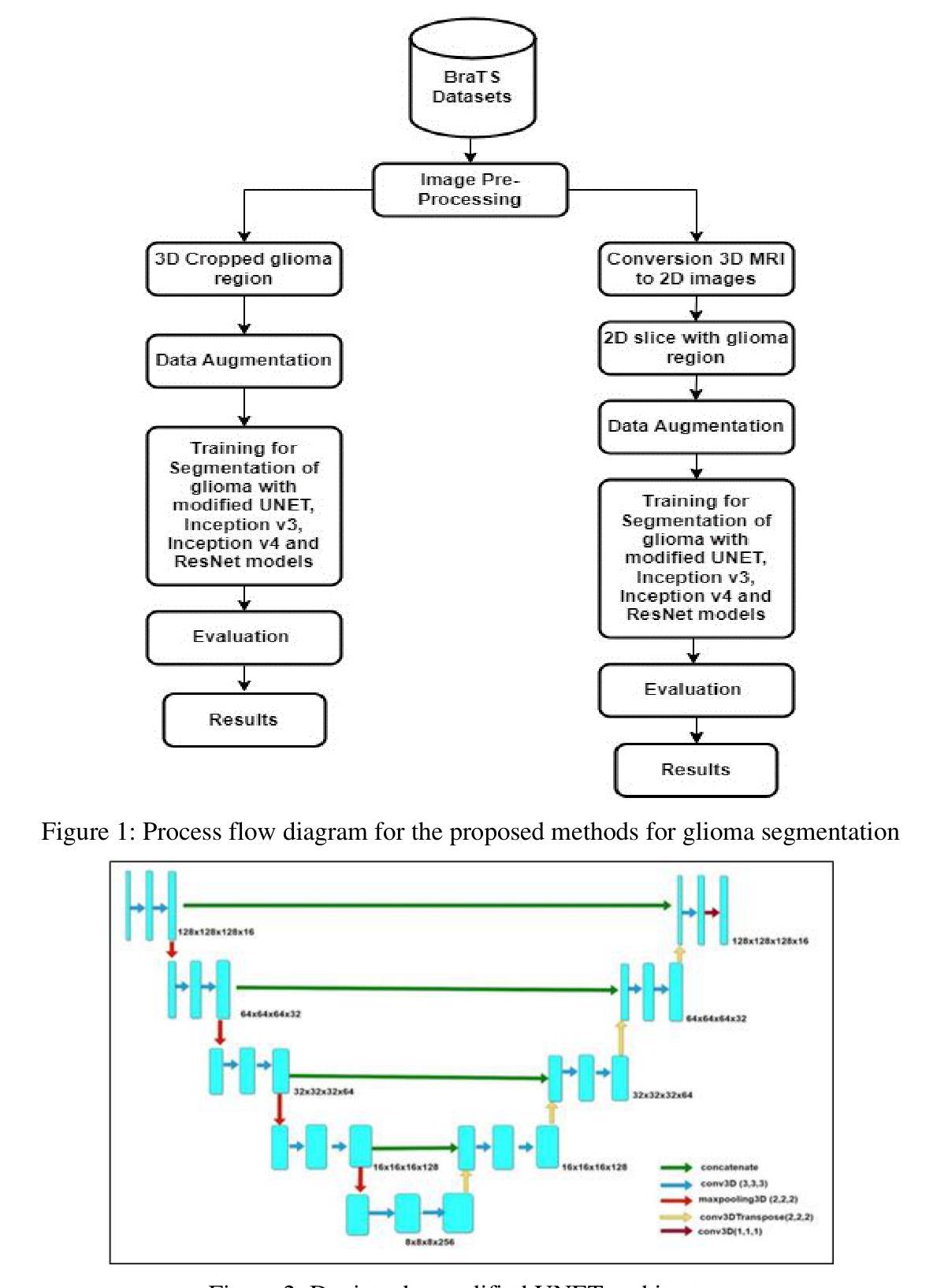
Brain tumor diagnosis is a challenging task for clinicians in the modern world. Among the major reasons for cancer-related death is the brain tumor. Gliomas, a category of central nervous system (CNS) tumors, encompass diverse subregions. For accurate diagnosis of brain tumors, precise segmentation of brain images and quantitative analysis are required. A fully automatic approach to glioma segmentation is required because the manual segmentation process is laborious, prone to mistakes, as well as time-consuming. Modern techniques for segmenting gliomas are based on fully convolutional neural networks (FCNs), which can either use two-dimensional (2D) or three-dimensional (3D) convolutions. Nevertheless, 3D convolutions suffer from computational costs and memory demand, while 2D convolutions cannot fully utilize the spatial insights of volumetric clinical imaging data. To obtain an optimal solution, it is vital to balance the computational efficiency of 2D convolutions along with the spatial accuracy of 3D convolutions. This balance can potentially be realized by developing an advanced model to overcome these challenges. The 2D and 3D models implemented here are based on UNET architecture, Inception, and ResNet models. The research work has been implemented on the BraTS 2018, 2019, and 2020 datasets. The best performer of all the models’ evaluations metrics for proposed methodologies offer superior potential in terms of the effective segmentation of gliomas. The ResNet model has resulted in 98.91% accuracy for 3D segmentation and 99.77 for 2D segmentations. The dice scores for 2D and 3D segmentations are 0.8312 and 0.9888, respectively. This model can be applied to various other medical applications with fine-tuning, thereby aiding clinicians in brain tumor analysis and improving the diagnosis process effectively.
脑肿瘤诊断是现代临床医生面临的一项具有挑战性的任务。脑肿瘤是导致癌症相关死亡的主要原因之一。胶质是中枢神经系统肿瘤的类别之一,涵盖多种亚区。为了准确诊断脑肿瘤,需要对脑图像进行精确分割和定量分析。由于手动分割过程既繁琐又容易出错且耗时,因此需要一种全自动的胶质瘤分割方法。现代用于分割胶质瘤的技术基于全卷积神经网络(FCNs),可以使用二维(2D)或三维(3D)卷积。然而,三维卷积存在计算成本高和内存需求大的问题,而二维卷积则无法充分利用体积临床成像数据的空间信息。为了获得最佳解决方案,平衡二维卷积的计算效率和三维卷积的空间精度至关重要。这种平衡可以通过开发一种先进的模型来克服这些挑战而实现。这里实现的二维和三维模型基于UNET架构、Inception和ResNet模型。研究工作已在BraTS 2018、2019和2020数据集上得到实施。所提出方法论的模型评估指标的最佳表现者提供了出色的潜力,在有效分割胶质瘤方面表现出卓越的性能。ResNet模型在三维分割方面达到了98.91%的准确率,在二维分割方面达到了99.77%。二维和三维分割的dice得分分别为0.8312和0.9888。通过微调,此模型可应用于其他各种医学应用,从而帮助临床医生进行脑肿瘤分析并有效改进诊断过程。
论文及项目相关链接
PDF 20 pages, 11 figures, journal paper
Summary
本文讨论了脑肿瘤诊断的挑战,特别是胶质瘤的精确分割和定量分析的重要性。由于手动分割过程繁琐、易出错且耗时,需要全自动的胶质瘤分割方法。当前技术基于全卷积神经网络(FCNs)的2D和3D卷积,但3D卷积计算成本高、内存需求大,而2D卷积不能充分利用体积临床影像数据的空间信息。研究平衡了2D卷积的计算效率和3D卷积的空间准确性,在BraTS数据集上实施,表现出优越的分割潜力,可有效辅助临床医生进行脑肿瘤分析和诊断。
Key Takeaways
- 脑肿瘤诊断在现代医学中仍是一项挑战,需要精确的图像分割和定量分析。
- 胶质瘤是中枢神经系统肿瘤的一种,其准确诊断需要全自动的分割方法。
- 当前技术使用基于全卷积神经网络(FCNs)的2D和3D卷积进行分割。
- 3D卷积存在计算成本高和内存需求大的问题,而2D卷积无法充分利用体积临床影像数据的空间信息。
- 研究旨在平衡计算效率和空间准确性,通过开发先进模型克服这些挑战。
- 使用UNET架构、Inception和ResNet模型的2D和3D模型在BraTS数据集上实施,表现优越。
点此查看论文截图

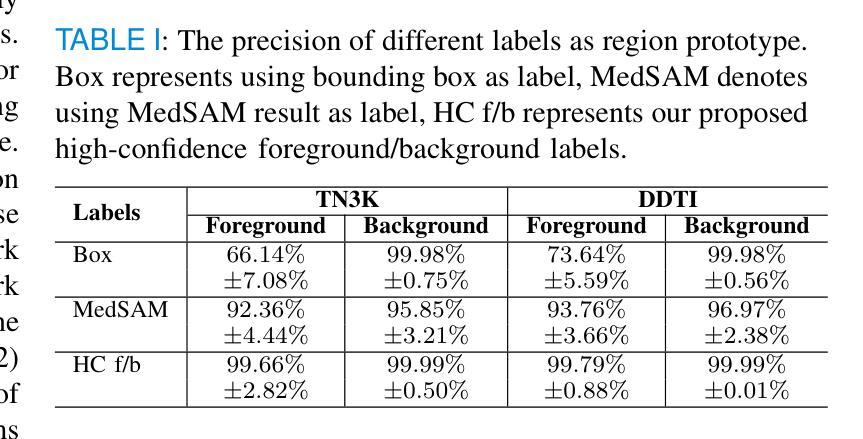
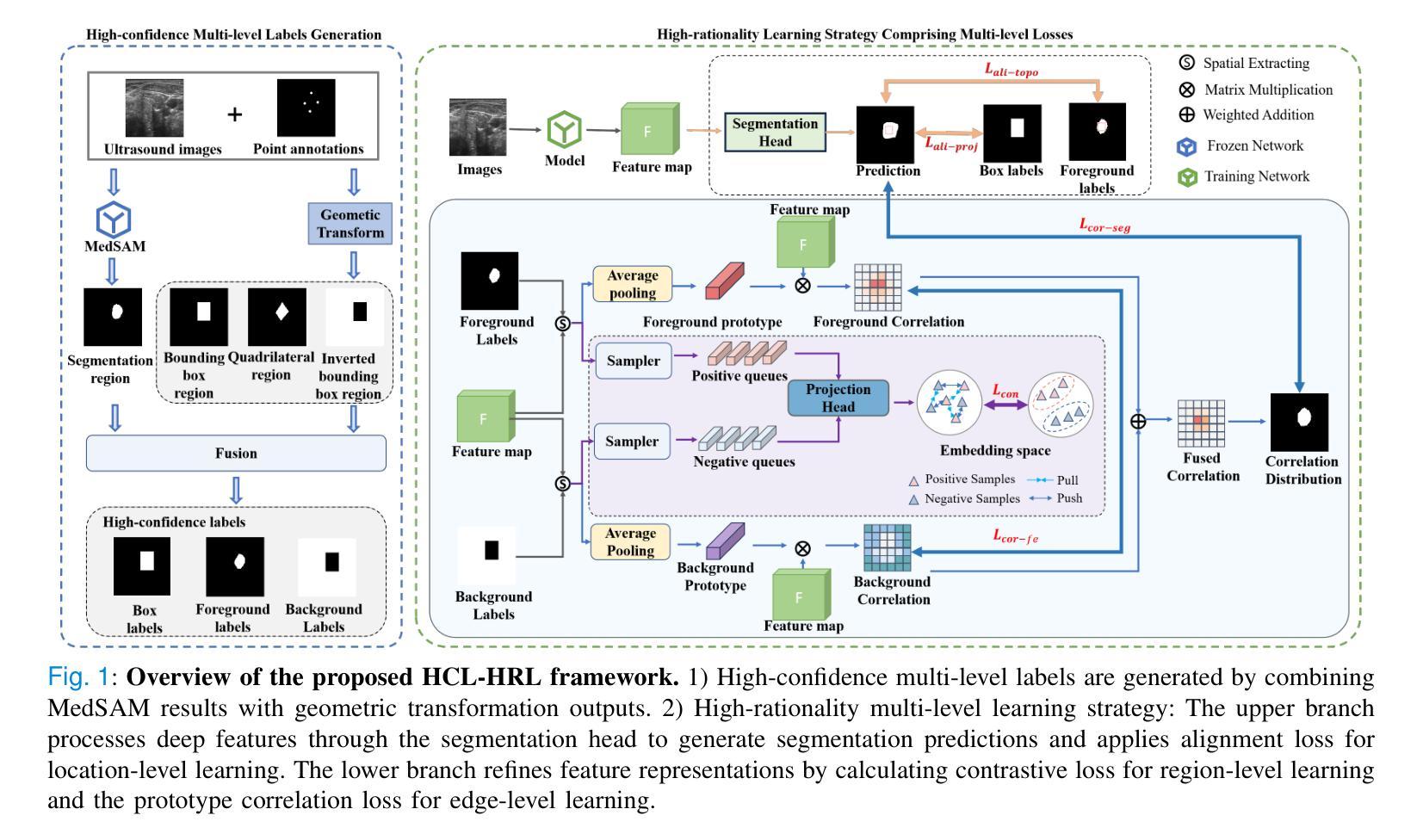
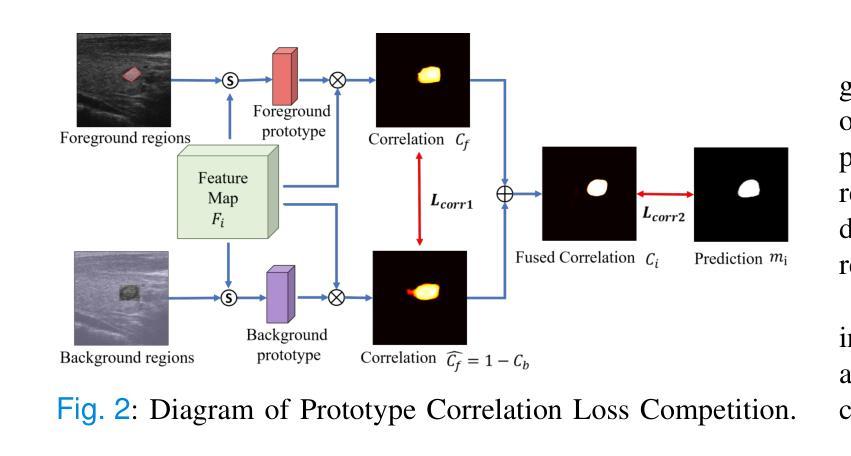
Weakly Supervised Segmentation Framework for Thyroid Nodule Based on High-confidence Labels and High-rationality Losses
Authors:Jianning Chi, Zelan Li, Geng Lin, MingYang Sun, Xiaosheng Yu
Weakly supervised segmentation methods can delineate thyroid nodules in ultrasound images efficiently using training data with coarse labels, but suffer from: 1) low-confidence pseudo-labels that follow topological priors, introducing significant label noise, and 2) low-rationality loss functions that rigidly compare segmentation with labels, ignoring discriminative information for nodules with diverse and complex shapes. To solve these issues, we clarify the objective and references for weakly supervised ultrasound image segmentation, presenting a framework with high-confidence pseudo-labels to represent topological and anatomical information and high-rationality losses to capture multi-level discriminative features. Specifically, we fuse geometric transformations of four-point annotations and MedSAM model results prompted by specific annotations to generate high-confidence box, foreground, and background labels. Our high-rationality learning strategy includes: 1) Alignment loss measuring spatial consistency between segmentation and box label, and topological continuity within the foreground label, guiding the network to perceive nodule location; 2) Contrastive loss pulling features from labeled foreground regions while pushing features from labeled foreground and background regions, guiding the network to learn nodule and background feature distribution; 3) Prototype correlation loss measuring consistency between correlation maps derived by comparing features with foreground and background prototypes, refining uncertain regions to accurate nodule edges. Experimental results show that our method achieves state-of-the-art performance on the TN3K and DDTI datasets. The code is available at https://github.com/bluehenglee/MLI-MSC.
弱监督分割方法能够利用带有粗标签的训练数据有效地对超声图像中的甲状腺结节进行描绘,但存在以下问题:1)遵循拓扑先验的低置信度伪标签,引入了大量的标签噪声;2)损失函数缺乏理性,僵化地将分割结果与标签进行比较,忽略了具有多样性和复杂形状的结节的判别信息。为了解决这些问题,我们明确了弱监督超声图像分割的目标和参考,提出了一个框架,采用高置信度伪标签来表示拓扑和解剖信息,以及高理性损失来捕捉多级别判别特征。具体来说,我们融合了四点注释的几何变换和MedSAM模型结果,生成了高置信度的框、前景和背景标签。我们的高理性学习策略包括:1)对齐损失,测量分割与框标签之间的空间一致性,以及前景标签内的拓扑连续性,指导网络感知结节位置;2)对比损失,从标记的前景区域中提取特征,同时从标记的前景和背景区域中推挽特征,指导网络学习结节和背景特征分布;3)原型关联损失,测量通过比较特征与前景和背景原型得到的关联图之间的一致性,精确修正不确定区域以得到准确的结节边缘。实验结果表明,我们的方法在TN3K和DDTI数据集上达到了最新性能。代码可在https://github.com/bluehenglee/MLI-MSC找到。
论文及项目相关链接
PDF 10 pages, 6 figures
Summary
本文提出一种基于弱监督学习的甲状腺超声图像分割方法,解决了现有方法存在的标签噪声和低理性损失函数问题。通过融合四点标注的几何变换和MedSAM模型结果,生成高置信度的标签。同时,引入高理性学习策略,包括对齐损失、对比损失和原型关联损失,以捕捉多层次特征。实验结果显示,该方法在TN3K和DDTI数据集上达到领先水平。
Key Takeaways
- 弱监督分割方法能高效利用粗标签数据对甲状腺结节进行超声图像分割。
- 现有方法存在标签噪声问题,主要是由于低置信度的伪标签和拓扑先验导致的。
- 融合四点标注的几何变换和MedSAM模型结果生成高置信度标签。
- 提出高理性学习策略,包括对齐损失、对比损失和原型关联损失,以捕捉多层次特征并优化不确定区域。
- 对齐损失衡量分割与框选标签的空间一致性及前景标签的拓扑连续性。
- 对比损失拉近前景区域的特征,同时推开前景和背景区域的特征,使网络学习结节和背景的特征分布。
- 原型关联损失衡量特征与前背景原型间的关联一致性,以优化不确定区域至准确的结节边缘。
点此查看论文截图
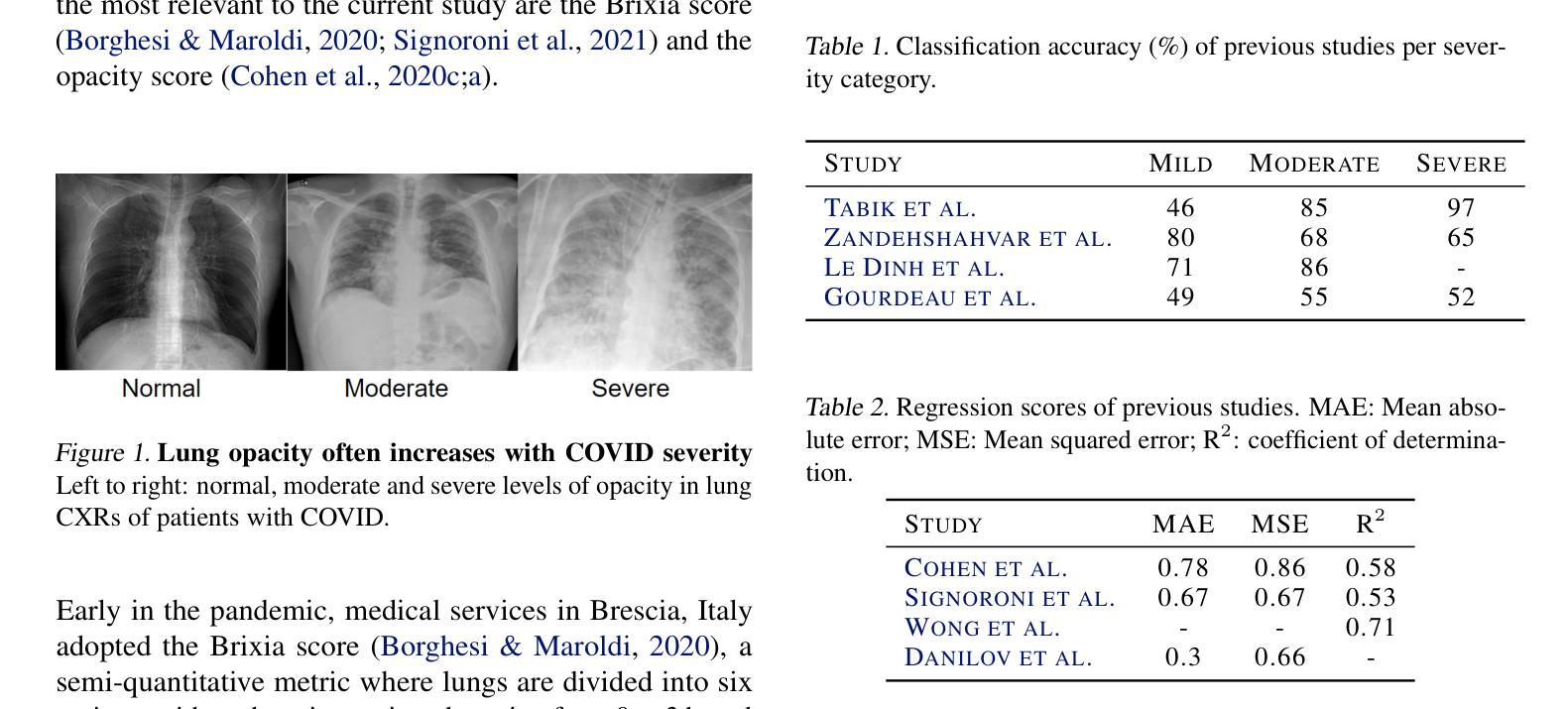
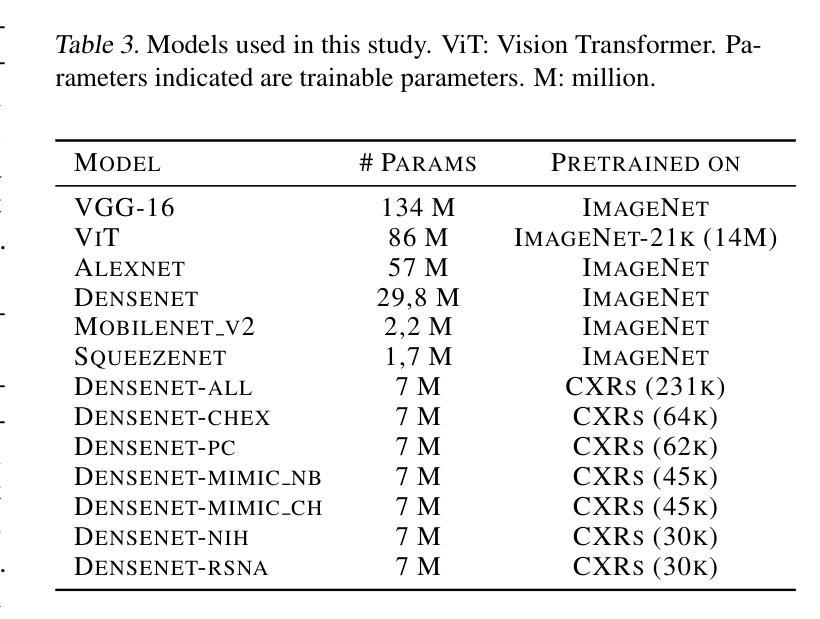
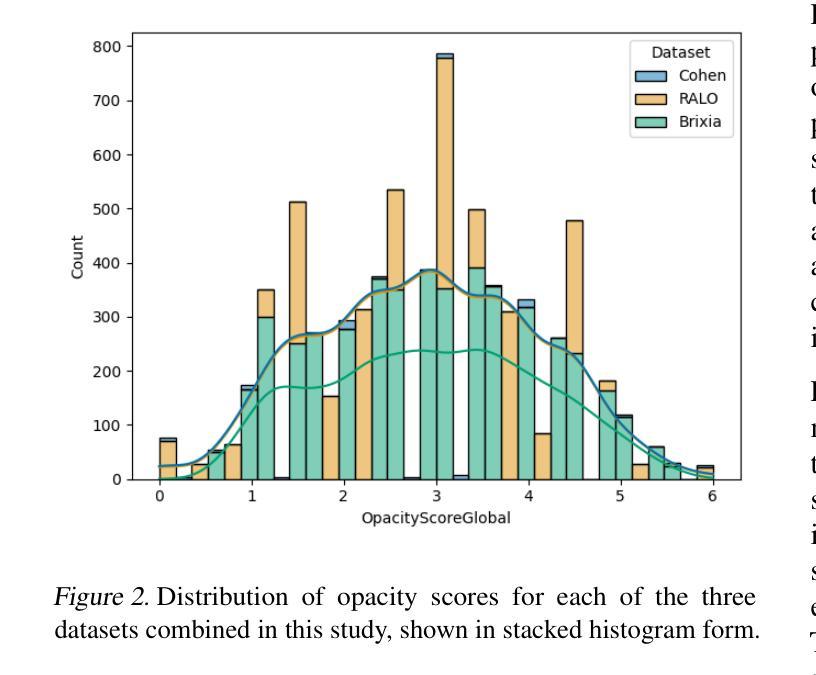
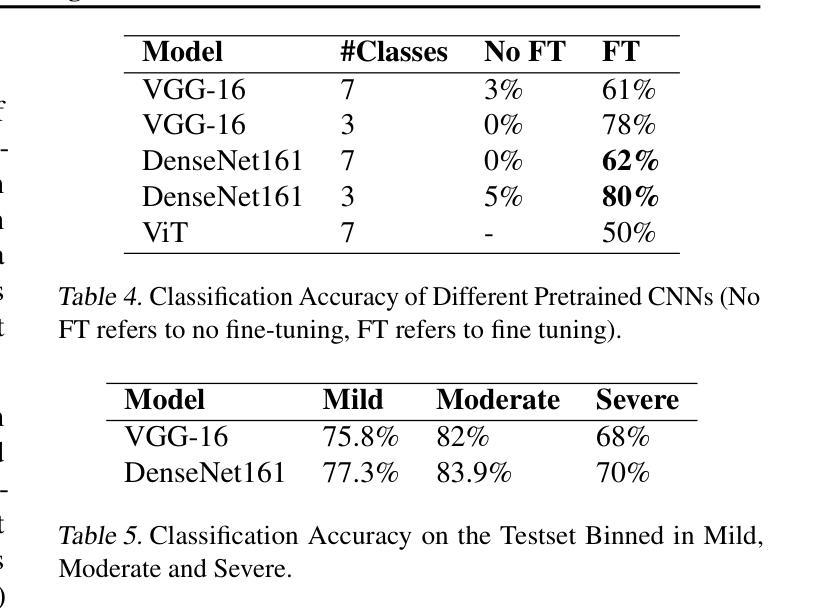
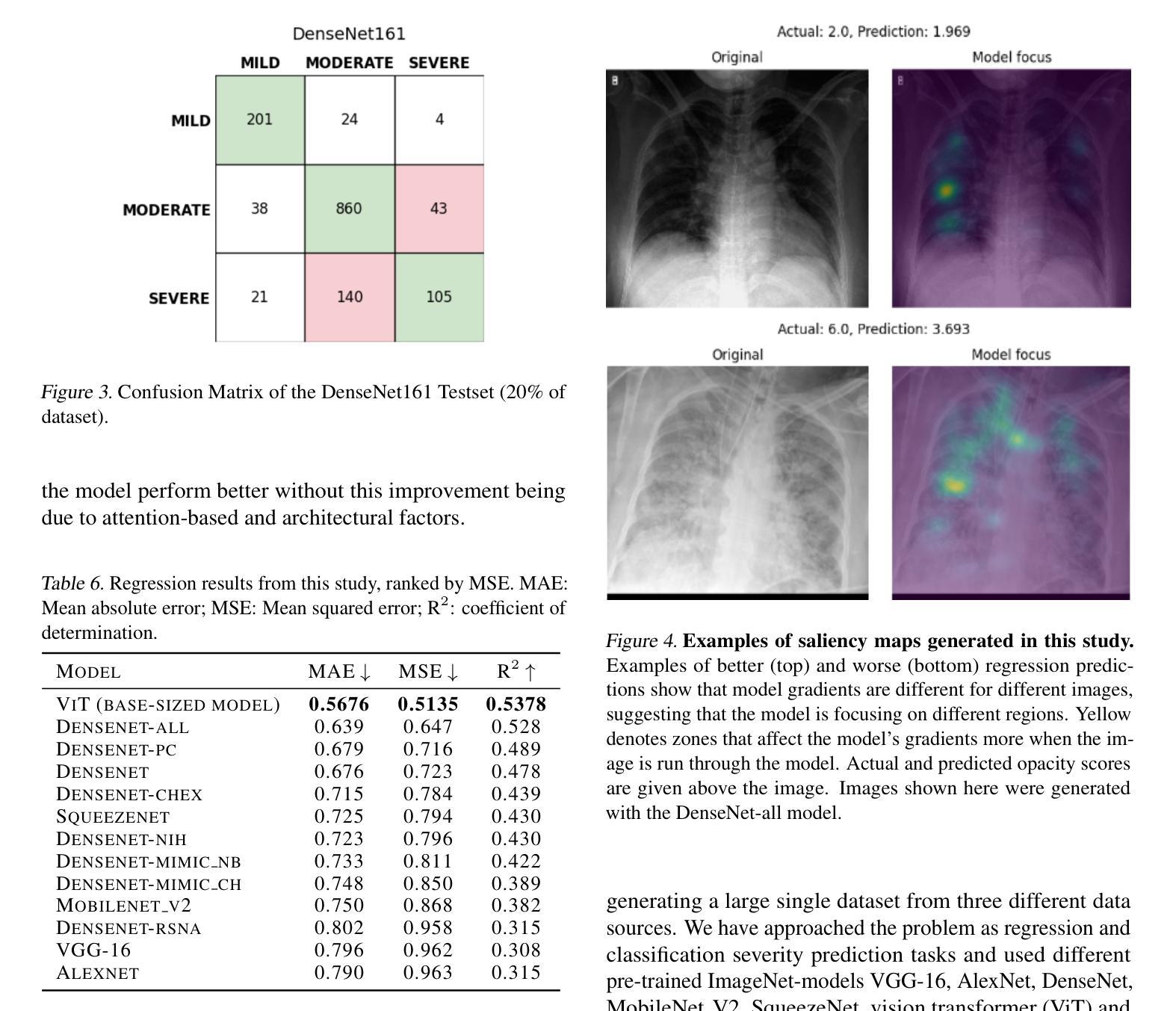
Diagnosing COVID-19 Severity from Chest X-Ray Images Using ViT and CNN Architectures
Authors:Luis Lara, Lucia Eve Berger, Rajesh Raju
The COVID-19 pandemic strained healthcare resources and prompted discussion about how machine learning can alleviate physician burdens and contribute to diagnosis. Chest x-rays (CXRs) are used for diagnosis of COVID-19, but few studies predict the severity of a patient’s condition from CXRs. In this study, we produce a large COVID severity dataset by merging three sources and investigate the efficacy of transfer learning using ImageNet- and CXR-pretrained models and vision transformers (ViTs) in both severity regression and classification tasks. A pretrained DenseNet161 model performed the best on the three class severity prediction problem, reaching 80% accuracy overall and 77.3%, 83.9%, and 70% on mild, moderate and severe cases, respectively. The ViT had the best regression results, with a mean absolute error of 0.5676 compared to radiologist-predicted severity scores. The project’s source code is publicly available.
COVID-19大流行使医疗资源承受巨大压力,并引发了关于机器学习如何减轻医生负担并为诊断做出贡献的讨论。胸部X射线(CXRs)被用于诊断COVID-19,但很少有研究从CXRs预测患者病情的严重程度。在本研究中,我们通过合并三个来源来创建了一个大规模的COVID严重程度数据集,并探讨了使用ImageNet和CXR预训练模型以及视觉转换器(ViTs)进行迁移学习在严重程度回归和分类任务中的有效性。在三级严重程度预测问题上,预训练的DenseNet161模型表现最佳,总体准确率达到80%,对轻度、中度和重度病例的准确率分别为77.3%、83.9%和70%。ViT在回归方面表现最佳,与放射科医生预测的严重程度评分相比,平均绝对误差为0.5676。该项目的源代码公开可用。
论文及项目相关链接
Summary
本文探讨了新冠肺炎疫情下医疗资源紧张的问题,以及机器学习如何缓解医生负担并助力诊断。研究通过合并三个来源的数据集,创建了一个大型的COVID-19病情严重程度数据集。同时,该研究还探究了使用ImageNet和CXR预训练模型以及视觉转换器(ViTs)进行迁移学习的效果,在病情严重程度回归和分类任务中表现良好。其中,预训练的DenseNet161模型在三类病情预测问题上表现最佳,总体准确率为80%,对轻度、中度和重度病例的准确率分别为77.3%、83.9%和70%。而ViT在回归任务中的表现最佳,与放射科医生预测的严重程度分数的平均绝对误差为0.5676。该项目的源代码已公开。
Key Takeaways
- 研究探讨了机器学习在缓解医生负担和助力COVID-19诊断方面的作用。
- 通过合并三个来源的数据集,创建了一个大型的COVID-19病情严重程度数据集。
- 使用了预训练的DenseNet161模型和视觉转换器(ViTs)进行迁移学习。
- DenseNet161模型在三类病情预测问题上表现最佳,总体准确率为80%。
- ViT在回归任务中的表现最佳,与放射科医生预测的平均绝对误差为0.5676。
- 该研究强调了机器学习和人工智能在医疗诊断中的潜力。
点此查看论文截图

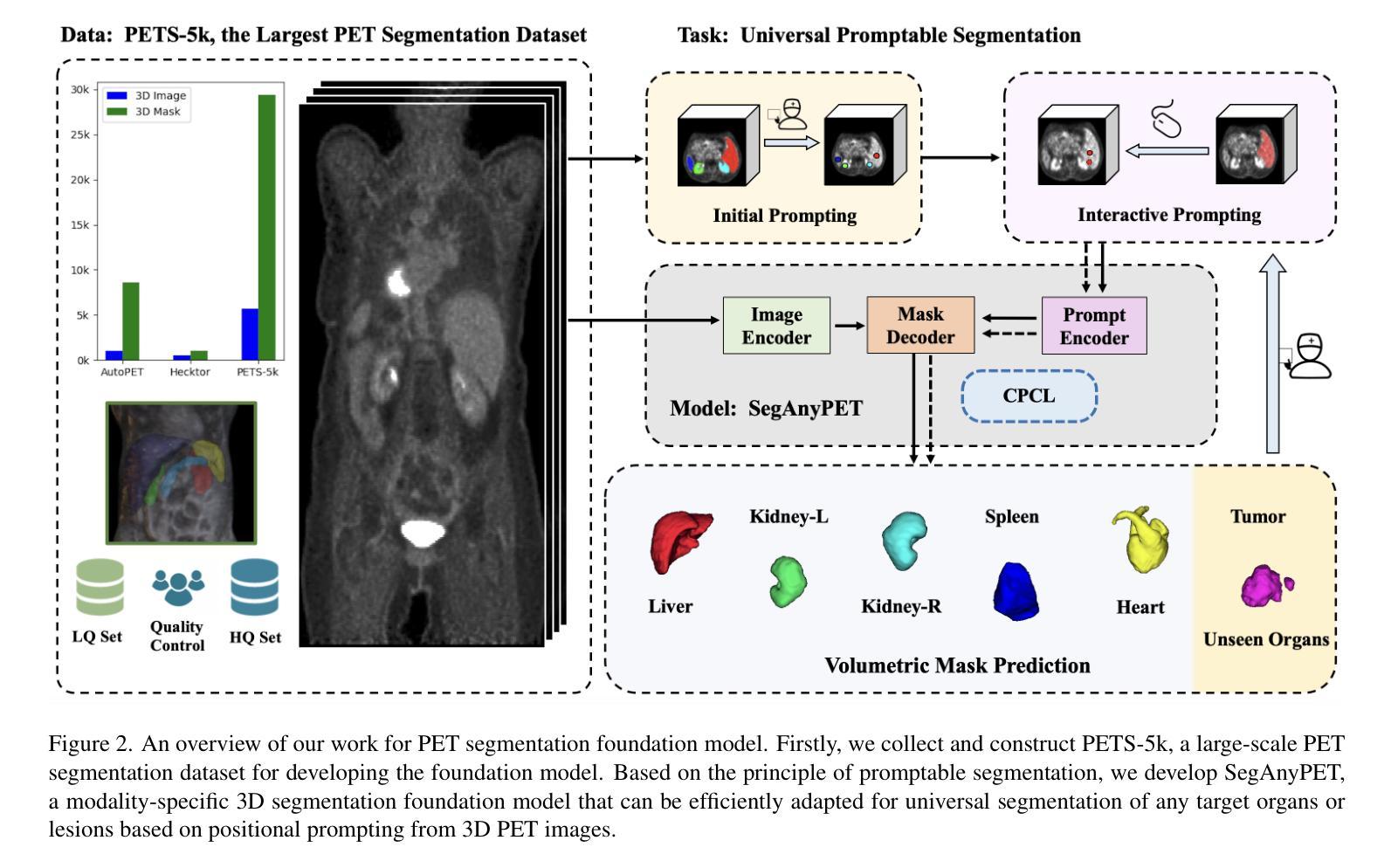
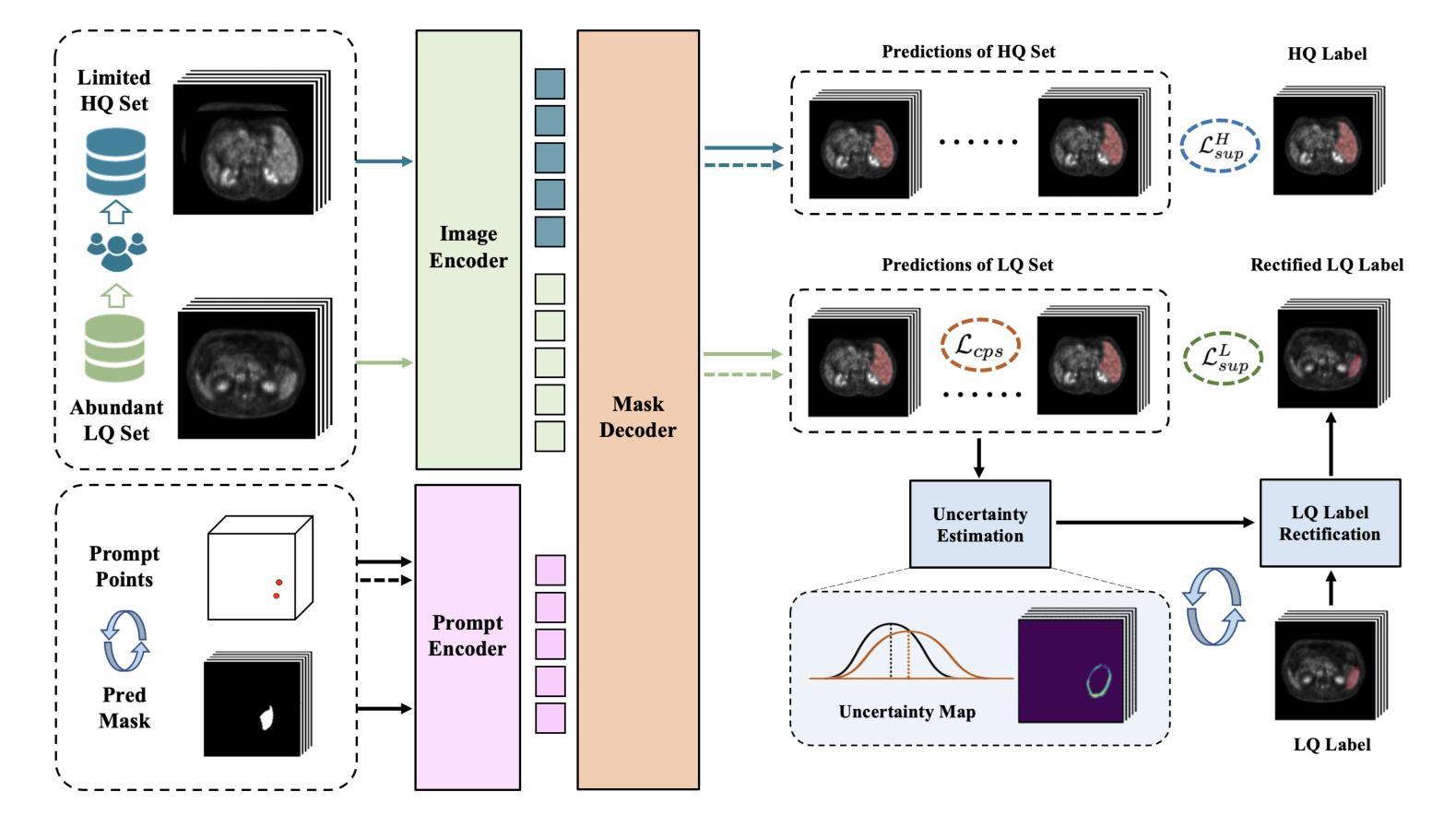
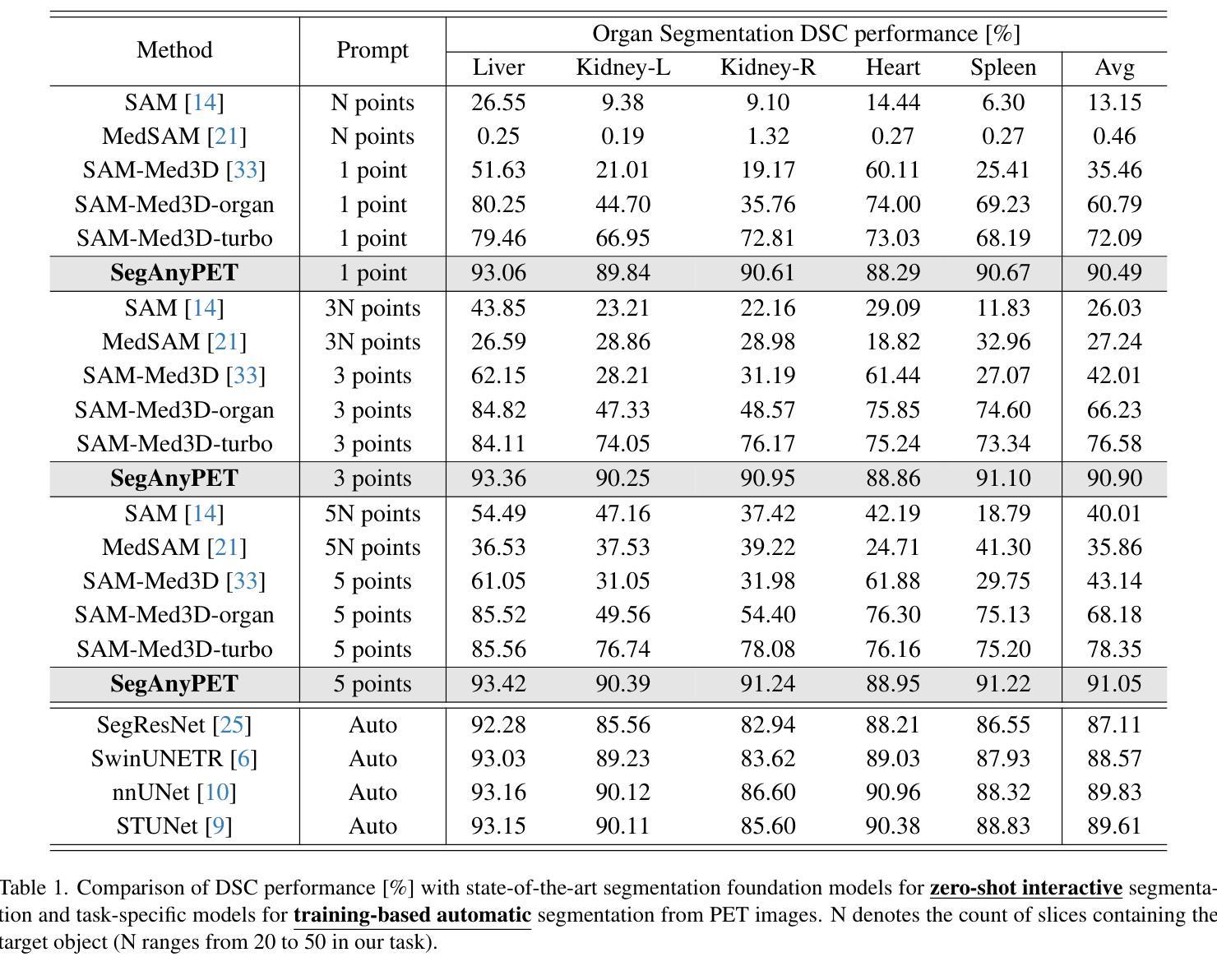
SegAnyPET: Universal Promptable Segmentation from Positron Emission Tomography Images
Authors:Yichi Zhang, Le Xue, Wenbo Zhang, Lanlan Li, Yuchen Liu, Chen Jiang, Yuan Cheng, Yuan Qi
Positron Emission Tomography (PET) imaging plays a crucial role in modern medical diagnostics by revealing the metabolic processes within a patient’s body, which is essential for quantification of therapy response and monitoring treatment progress. However, the segmentation of PET images presents unique challenges due to their lower contrast and less distinct boundaries compared to other structural medical modalities. Recent developments in segmentation foundation models have shown superior versatility across diverse natural image segmentation tasks. Despite the efforts of medical adaptations, these works primarily focus on structural medical images with detailed physiological structural information and exhibit poor generalization ability when adapted to molecular PET imaging. In this paper, we collect and construct PETS-5k, the largest PET segmentation dataset to date, comprising 5,731 three-dimensional whole-body PET images and encompassing over 1.3M 2D images. Based on the established dataset, we develop SegAnyPET, a modality-specific 3D foundation model for universal promptable segmentation from PET images. To issue the challenge of discrepant annotation quality of PET images, we adopt a cross prompting confident learning (CPCL) strategy with an uncertainty-guided self-rectification process to robustly learn segmentation from high-quality labeled data and low-quality noisy labeled data. Experimental results demonstrate that SegAnyPET can correctly segment seen and unseen targets using only one or a few prompt points, outperforming state-of-the-art foundation models and task-specific fully supervised models with higher accuracy and strong generalization ability for universal segmentation. As the first foundation model for PET images, we believe that SegAnyPET will advance the applications to various downstream tasks for molecular imaging.
正电子发射断层扫描(PET)成像在现代医学诊断中发挥着关键作用,它能够揭示患者体内的代谢过程,这对于量化治疗反应和监测治疗进展至关重要。然而,由于PET图像的对比度较低,边界不够清晰,与其他结构医学图像相比,其分割面临独特挑战。最近的分割基础模型的发展表明,它们在各种自然图像分割任务中具有出色的多功能性。尽管进行了医学适应性的努力,但这些工作主要集中在具有详细生理结构信息的结构医学图像上,而在适应分子PET成像时表现出较差的泛化能力。在本文中,我们收集和构建了迄今为止最大的PET分割数据集PETS-5k,包含5731个三维全身PET图像和超过130万张二维图像。基于建立的数据集,我们开发了SegAnyPET,这是一个针对PET图像的特定模态的3D基础模型,用于通用的即时分割。针对PET图像标注质量不一致的问题,我们采用带有不确定性引导的自我修正过程的交叉提示置信学习(CPCL)策略,从高质量标注数据和低质量噪声标注数据中稳健地学习分割。实验结果表明,SegAnyPET仅使用一个或少数提示点就能正确分割已见和未见的目标,超越了最新的基础模型和任务特定的全监督模型,具有更高的准确性和强大的泛化能力,可用于通用的分割。作为第一个用于PET图像的基础模型,我们相信SegAnyPET将推动其在分子成像的各种下游任务中的应用。
论文及项目相关链接
Summary
本文介绍了正电子发射断层扫描(PET)成像在现代医学诊断中的重要性,并指出了PET图像分割面临的挑战。为此,作者构建了迄今为止最大的PET分割数据集PETS-5k,并基于此开发了SegAnyPET,一个用于PET图像通用分割的模态特定三维基础模型。为解决PET图像标注质量不一的问题,采用跨提示置信学习(CPCL)策略,结合不确定性引导的自我修正过程,从高质量和有噪声的标签数据中稳健地学习分割。实验结果表明,SegAnyPET能正确分割已见和未见目标,只用一两个提示点即可超越现有基础模型和任务特定全监督模型的性能,具有强大的通用分割能力。
Key Takeaways
- PET成像在现代医学诊断中揭示患者体内代谢过程,对疗效评估和追踪治疗进展至关重要。
- PET图像分割面临低对比度和边界模糊的挑战。
- PETS-5k数据集的建立解决了PET图像分割的瓶颈问题,是最大的PET分割数据集。
- SegAnyPET是首个针对PET图像的基础模型,用于通用分割。
- CPCL策略解决了PET图像标注质量不一的问题,提高了模型的稳健性。
- SegAnyPET能正确分割已见和未见目标,只用少数提示点即可实现高性能分割。
点此查看论文截图

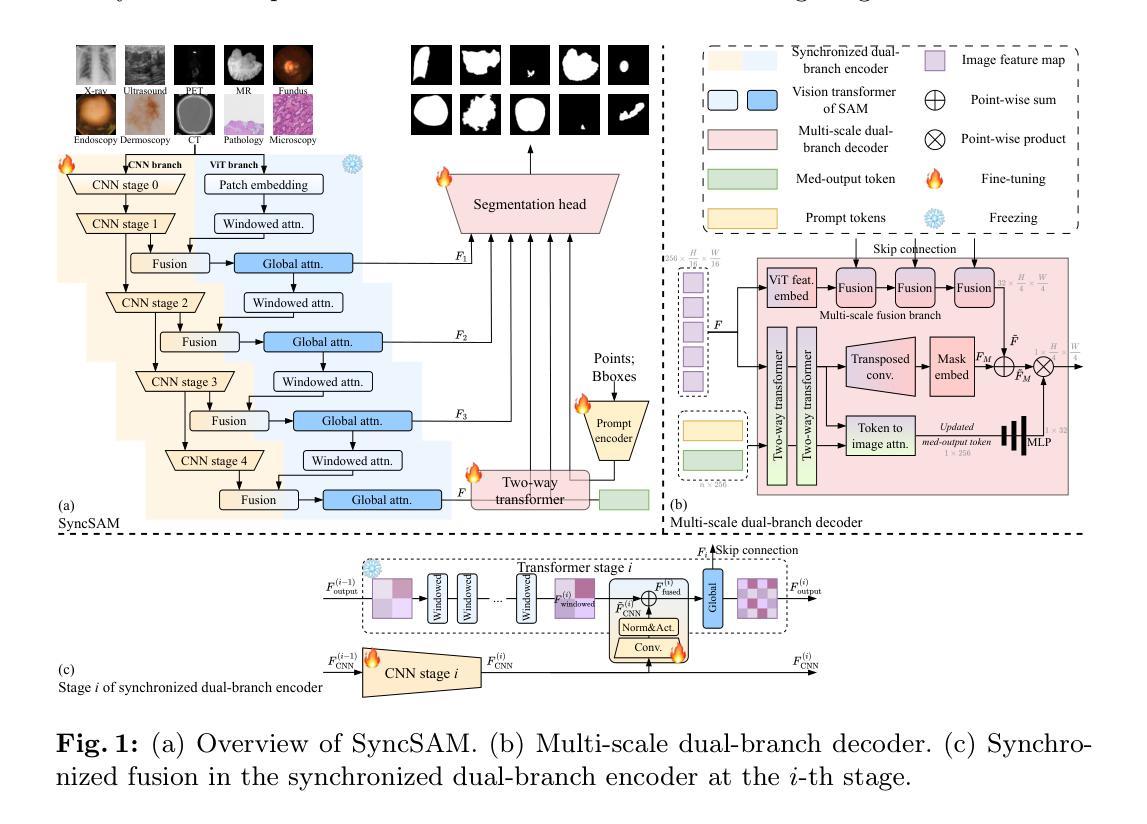
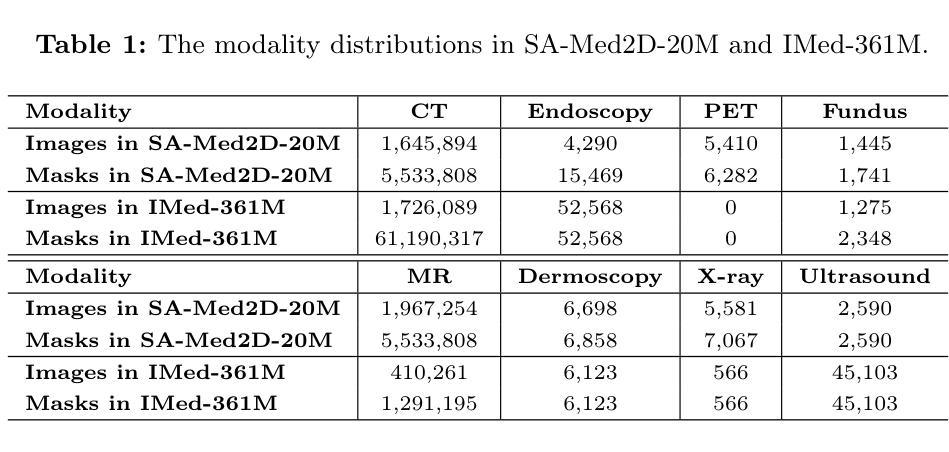
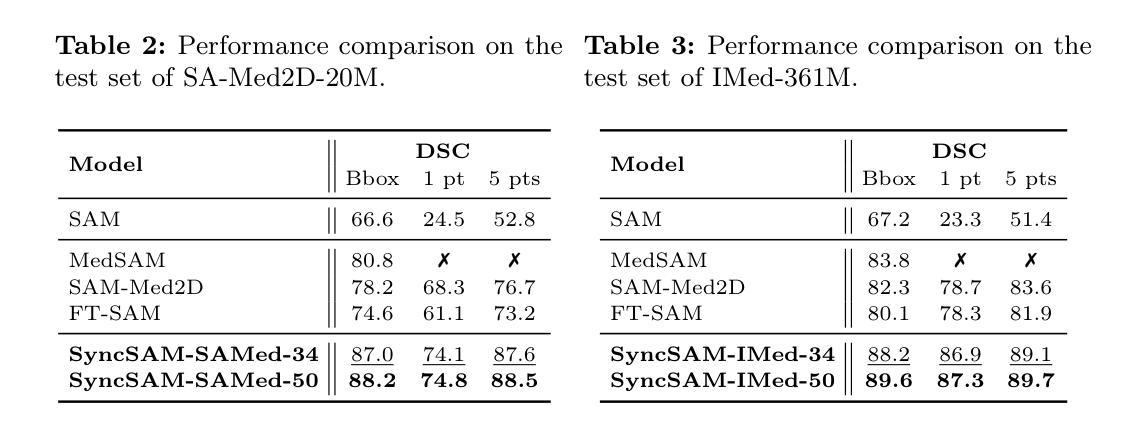
Improved Baselines with Synchronized Encoding for Universal Medical Image Segmentation
Authors:Sihan Yang, Xuande Mi, Jiadong Feng, Haixia Bi, Hai Zhang, Jian Sun
Large foundation models, known for their strong zero-shot generalization capabilities, can be applied to a wide range of downstream tasks. However, developing foundation models for medical image segmentation poses a significant challenge due to the domain gap between natural and medical images. While fine-tuning techniques based on the Segment Anything Model (SAM) have been explored, they primarily focus on scaling up data or refining inference strategies without incorporating domain-specific architectural designs, limiting their zero-shot performance. To optimize segmentation performance under standard inference settings and provide a strong baseline for future research, we introduce SyncSAM, which employs a synchronized dual-branch encoder that integrates convolution and Transformer features in a synchronized manner to enhance medical image encoding, and a multi-scale dual-branch decoder to preserve image details. SyncSAM is trained on two of the largest medical image segmentation datasets, SA-Med2D-20M and IMed-361M, resulting in a series of pre-trained models for universal medical image segmentation. Experimental results demonstrate that SyncSAM not only achieves state-of-the-art performance on test sets but also exhibits strong zero-shot capabilities on unseen datasets. The code and model weights are available at https://github.com/Hhankyangg/SyncSAM.
大型基础模型以其强大的零样本泛化能力而著称,可广泛应用于各种下游任务。然而,开发用于医学图像分割的基础模型构成了一项重大挑战,因为自然图像和医学图像之间存在领域差距。虽然基于Segment Anything Model(SAM)的微调技术已经被探索,但它们主要关注扩大数据规模或改进推理策略,而没有融入领域特定的架构设计,从而限制了其零样本性能。为了优化标准推理设置下的分割性能,并为未来的研究提供强大的基线,我们引入了SyncSAM。SyncSAM采用同步双分支编码器,以同步方式集成卷积和Transformer特征以增强医学图像编码,并采用多尺度双分支解码器以保留图像细节。SyncSAM在两大医学图像分割数据集SA-Med2D-20M和IMed-361M上进行训练,生成了一系列用于通用医学图像分割的预训练模型。实验结果表明,SyncSAM不仅在测试集上达到了最先进的性能,而且对未见数据集表现出了强大的零样本能力。相关代码和模型权重可在https://github.com/Hhankyangg/SyncSAM获取。
论文及项目相关链接
Summary
基于大型预训练模型在零样本学习方面的优势,本文提出了一种针对医学图像分割任务的同步双分支模型SyncSAM。该模型结合了卷积和Transformer特征,并采用了多尺度双分支解码器,旨在提高医学图像编码的效果并保留图像细节。实验结果表明,SyncSAM在测试集上达到了领先水平,并对未见数据集展现了强大的零样本学习能力。其预训练模型可用于通用医学图像分割任务,相关代码和模型权重已公开发布。
Key Takeaways
- 大型预训练模型在医学图像分割任务中具有潜力,但面临自然图像与医学图像领域差异的挑战。
- SyncSAM模型通过结合卷积和Transformer特征,增强医学图像编码效果。
- SyncSAM采用同步双分支编码器结构,旨在提高零样本学习能力。
- 模型在多个医学图像分割数据集上进行训练,包括SA-Med2D-20M和IMed-361M。
- 实验结果表明SyncSAM在测试集上表现优秀,达到领先水平。
- SyncSAM对未见数据集展现了强大的零样本学习能力。
点此查看论文截图

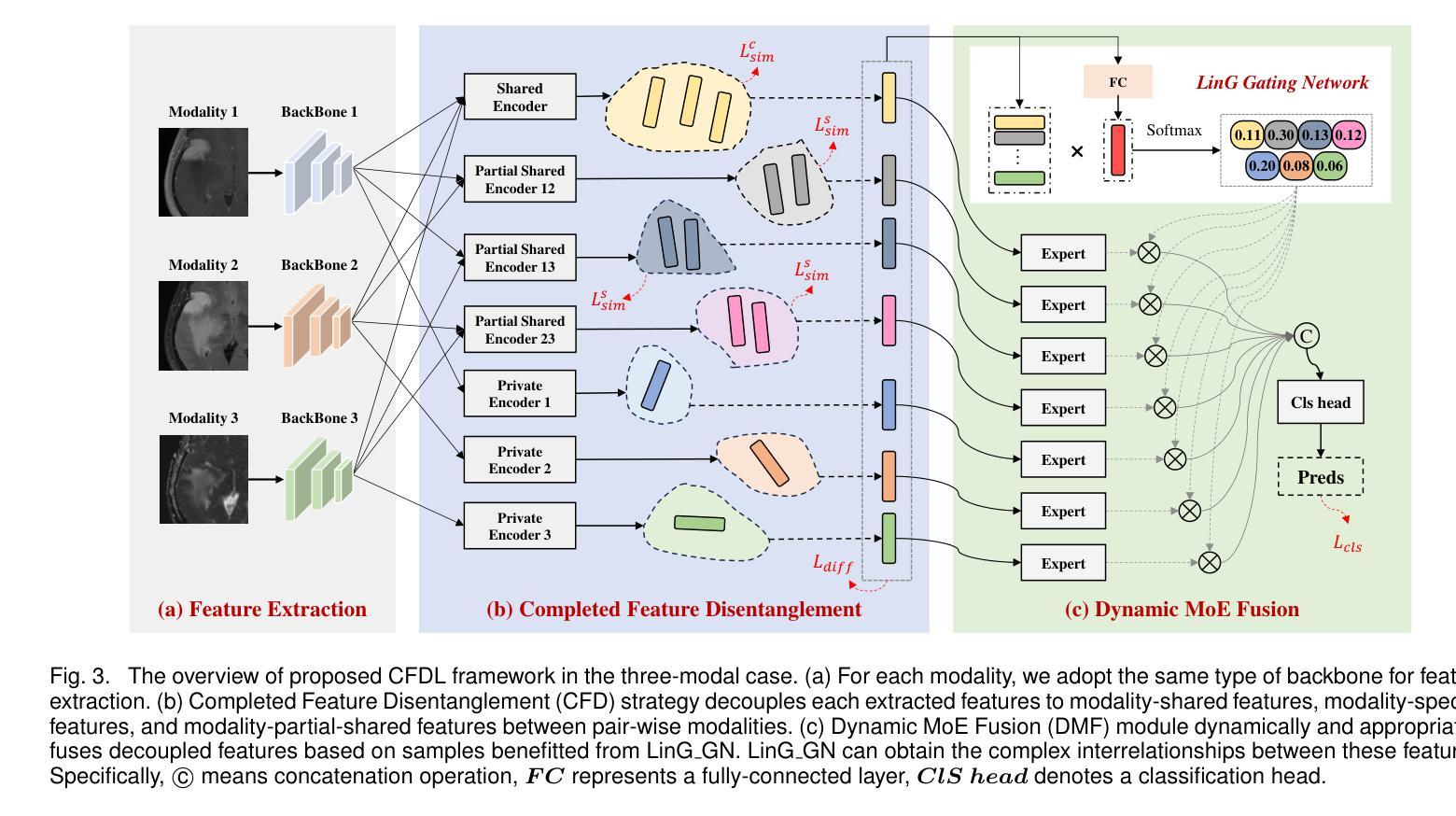
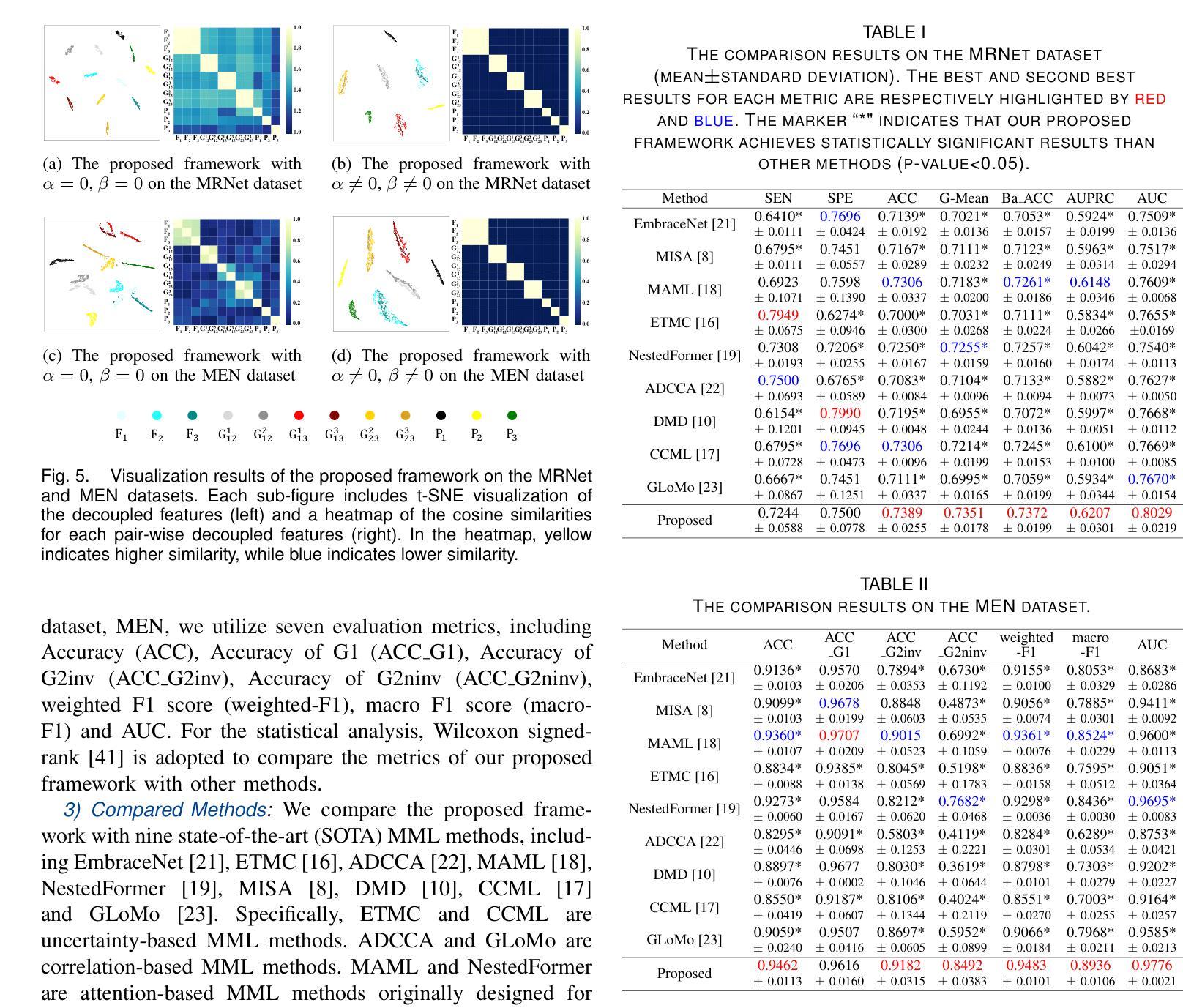
Completed Feature Disentanglement Learning for Multimodal MRIs Analysis
Authors:Tianling Liu, Hongying Liu, Fanhua Shang, Lequan Yu, Tong Han, Liang Wan
Multimodal MRIs play a crucial role in clinical diagnosis and treatment. Feature disentanglement (FD)-based methods, aiming at learning superior feature representations for multimodal data analysis, have achieved significant success in multimodal learning (MML). Typically, existing FD-based methods separate multimodal data into modality-shared and modality-specific features, and employ concatenation or attention mechanisms to integrate these features. However, our preliminary experiments indicate that these methods could lead to a loss of shared information among subsets of modalities when the inputs contain more than two modalities, and such information is critical for prediction accuracy. Furthermore, these methods do not adequately interpret the relationships between the decoupled features at the fusion stage. To address these limitations, we propose a novel Complete Feature Disentanglement (CFD) strategy that recovers the lost information during feature decoupling. Specifically, the CFD strategy not only identifies modality-shared and modality-specific features, but also decouples shared features among subsets of multimodal inputs, termed as modality-partial-shared features. We further introduce a new Dynamic Mixture-of-Experts Fusion (DMF) module that dynamically integrates these decoupled features, by explicitly learning the local-global relationships among the features. The effectiveness of our approach is validated through classification tasks on three multimodal MRI datasets. Extensive experimental results demonstrate that our approach outperforms other state-of-the-art MML methods with obvious margins, showcasing its superior performance.
多模态MRI在临床诊断和治疗中扮演着至关重要的角色。特征解构(FD)方法旨在为多模态数据分析学习优质特征表示,已在多模态学习(MML)中取得了显著的成功。通常,现有的FD方法将多模态数据分为模态共享和模态特定特征,并使用拼接或注意力机制来整合这些特征。然而,我们的初步实验表明,当输入包含超过两种模态时,这些方法可能会导致模态子集之间共享信息的丢失,且此类信息对预测精度至关重要。此外,这些方法在融合阶段没有充分解释解耦特征之间的关系。为了解决这些局限性,我们提出了一种新颖的特征解构策略,即完全特征解构(CFD),该策略恢复了特征解构过程中丢失的信息。具体来说,CFD策略不仅识别模态共享和模态特定特征,还解耦多模态输入子集之间的共享特征,称为模态部分共享特征。我们进一步引入了一个新的动态混合专家融合(DMF)模块,它通过显式学习特征之间的局部全局关系来动态整合这些解耦的特征。我们的方法通过三个多模态MRI数据集上的分类任务进行了验证。大量的实验结果表明,我们的方法在其他先进MML方法上优势明显,展示了其卓越性能。
论文及项目相关链接
PDF Accept by IEEE JBHI 2025
Summary
本文介绍了多模态MRI在临床诊断和治疗中的重要性,以及特征分解(FD)方法在多模态学习(MML)中的应用。现有FD方法将多模态数据分为共享和特定特征,并通过拼接或注意力机制进行集成。然而,当输入包含超过两种模态时,这些方法可能会丢失部分模态间的共享信息。为解决这个问题,提出了一种全新的完整特征分解(CFD)策略,能够恢复在特征分解过程中丢失的信息。此外,还引入了动态混合专家融合(DMF)模块,能够动态地融合这些分解后的特征,并显式地学习特征之间的局部-全局关系。通过在三组多模态MRI数据集上的分类任务验证,该方法显著优于其他最先进的MML方法。
Key Takeaways
- 多模态MRI在临床诊断和治疗中具有重要作用。
- 特征分解(FD)方法在多模态学习(MML)中取得了显著成功。
- 现有FD方法在处理超过两种模态的输入时,可能会丢失部分模态间的共享信息。
- 提出的完整特征分解(CFD)策略能够恢复在特征分解过程中丢失的信息。
- 引入了动态混合专家融合(DMF)模块,能动态融合特征并学习特征间的局部-全局关系。
- 该方法在三个多模态MRI数据集上的分类任务中表现出卓越性能。
点此查看论文截图