⚠️ 以下所有内容总结都来自于 大语言模型的能力,如有错误,仅供参考,谨慎使用
🔴 请注意:千万不要用于严肃的学术场景,只能用于论文阅读前的初筛!
💗 如果您觉得我们的项目对您有帮助 ChatPaperFree ,还请您给我们一些鼓励!⭐️ HuggingFace免费体验
2025-03-10 更新
RadIR: A Scalable Framework for Multi-Grained Medical Image Retrieval via Radiology Report Mining
Authors:Tengfei Zhang, Ziheng Zhao, Chaoyi Wu, Xiao Zhou, Ya Zhang, Yangfeng Wang, Weidi Xie
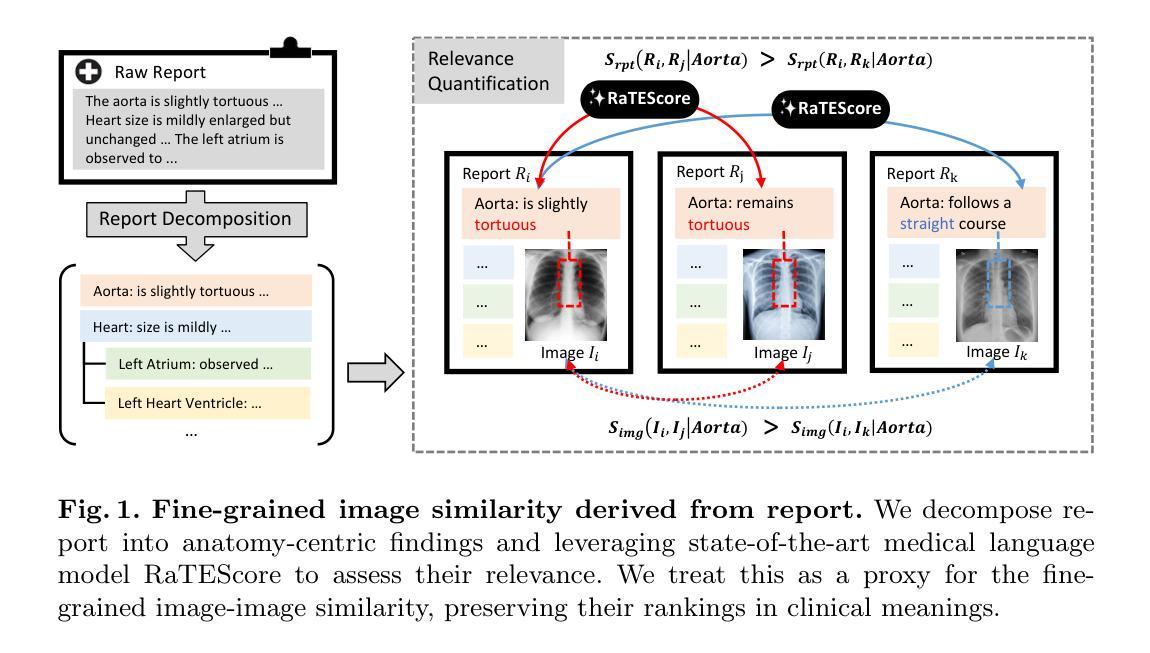
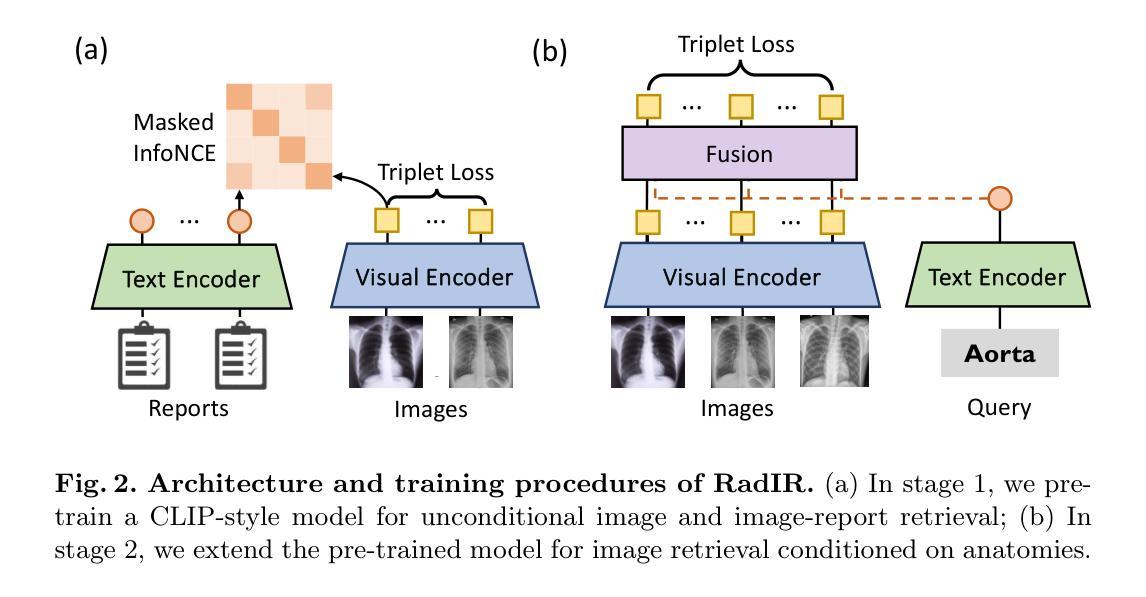
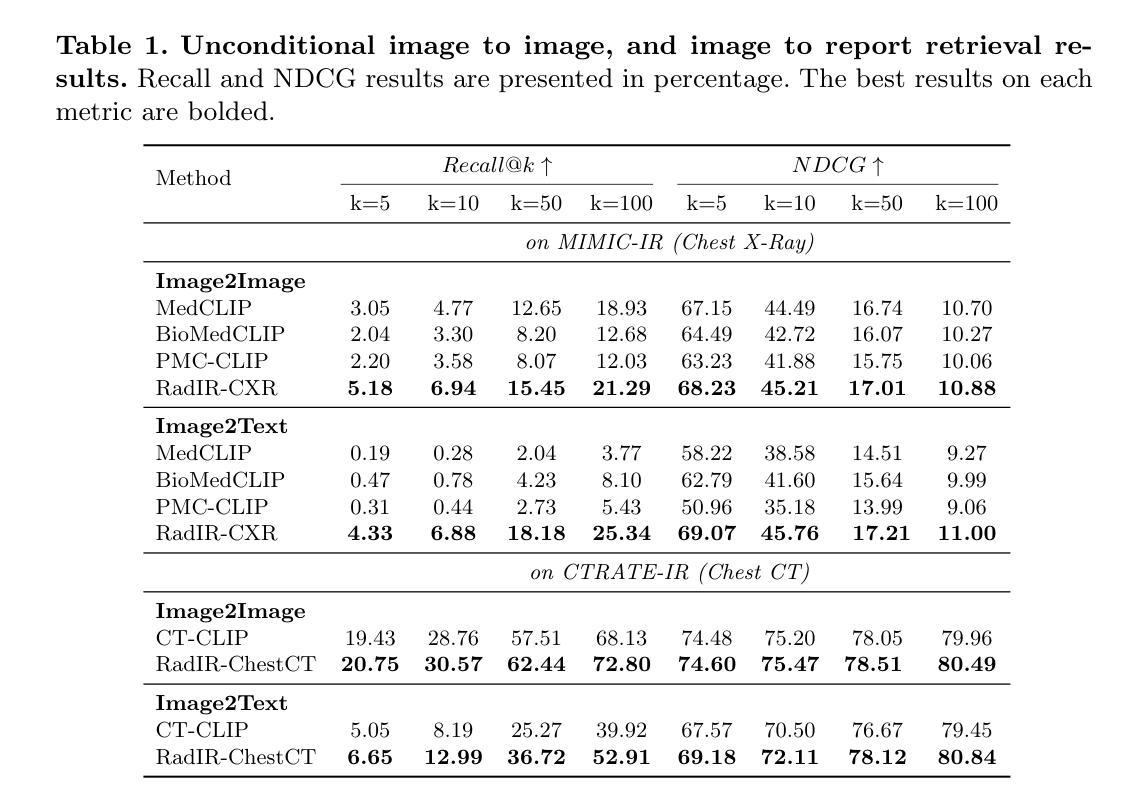
Developing advanced medical imaging retrieval systems is challenging due to the varying definitions of `similar images’ across different medical contexts. This challenge is compounded by the lack of large-scale, high-quality medical imaging retrieval datasets and benchmarks. In this paper, we propose a novel methodology that leverages dense radiology reports to define image-wise similarity ordering at multiple granularities in a scalable and fully automatic manner. Using this approach, we construct two comprehensive medical imaging retrieval datasets: MIMIC-IR for Chest X-rays and CTRATE-IR for CT scans, providing detailed image-image ranking annotations conditioned on diverse anatomical structures. Furthermore, we develop two retrieval systems, RadIR-CXR and model-ChestCT, which demonstrate superior performance in traditional image-image and image-report retrieval tasks. These systems also enable flexible, effective image retrieval conditioned on specific anatomical structures described in text, achieving state-of-the-art results on 77 out of 78 metrics.
开发先进的医学图像检索系统是一个挑战,因为“相似图像”的定义在不同的医学语境中各不相同。这一挑战由于缺乏大规模、高质量的医学图像检索数据集和基准测试而加剧。在本文中,我们提出了一种新方法,该方法利用密集的放射学报告来定义图像层面的相似性排序,以可伸缩和全自动的方式在多个粒度级别上实现。通过这种方法,我们构建了两个全面的医学图像检索数据集:用于胸部X射线的MIMIC-IR和用于CT扫描的CTRATE-IR,它们根据多种解剖结构提供了详细的图像-图像排名注释。此外,我们开发了两个检索系统RadIR-CXR和model-ChestCT,在传统的图像-图像和图像-报告检索任务中表现出卓越的性能。这些系统还能够根据文本中描述的特定解剖结构进行灵活有效的图像检索,在78个指标中的77个上取得了最先进的成果。
论文及项目相关链接
摘要
医学图像检索系统面临多种挑战,其中包括在不同医学语境下对“相似图像”定义的差异、缺乏大规模高质量医学图像检索数据集和基准测试等。本文提出了一种新的方法,通过密集的放射学报告来定义图像级相似性排序的多种粒度,并以可伸缩和全自动的方式进行。基于此方法,我们构建了两个全面的医学图像检索数据集:MIMIC-IR用于胸部X光检查和CTRATE-IR用于CT扫描,为基于不同解剖结构的图像图像排名提供了详细的注释。此外,我们开发了两个检索系统RadIR-CXR和model-ChestCT,在传统的图像图像和图像报告检索任务中表现出卓越的性能。这些系统还可以根据文本中描述的特定解剖结构实现灵活有效的图像检索,在78个指标中的77个上取得了最先进的成果。
关键见解
- 医学图像检索系统面临定义“相似图像”的挑战,这一挑战因缺乏大规模高质量数据集和基准测试而加剧。
- 提出了一种新的方法,利用密集的放射学报告来定义图像级相似性排序的多种粒度,并全自动执行。
- 构建了两个全面的医学图像检索数据集:MIMIC-IR和CTRATE-IR,为基于不同解剖结构的图像提供详细排名注释。
- 开发了两个高性能的医学图像检索系统:RadIR-CXR和model-ChestCT。
- 这些系统可以灵活有效地根据文本描述的特定解剖结构进行图像检索。
- 在大部分评估指标上,这些系统达到了最先进的表现。
点此查看论文截图

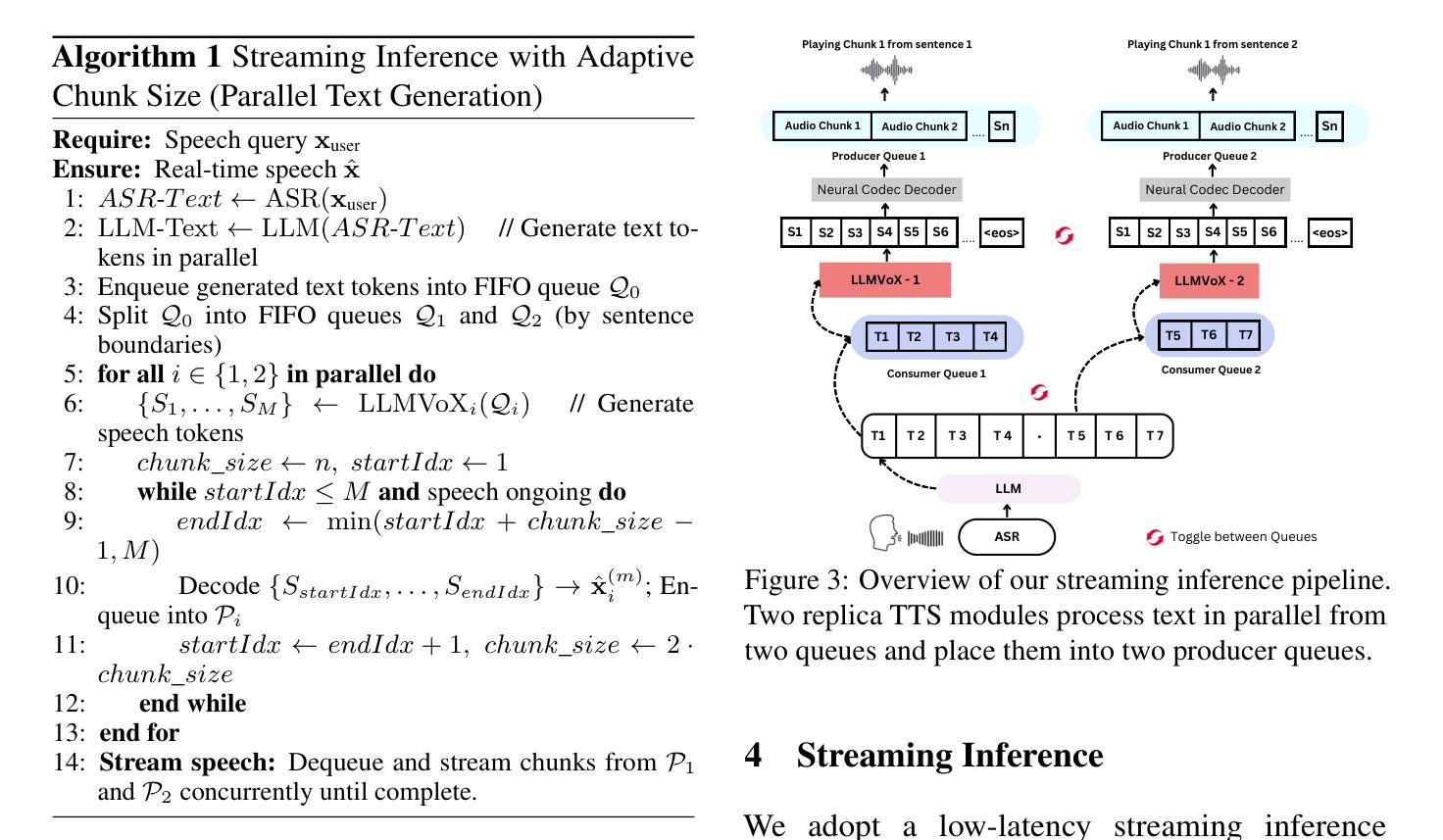
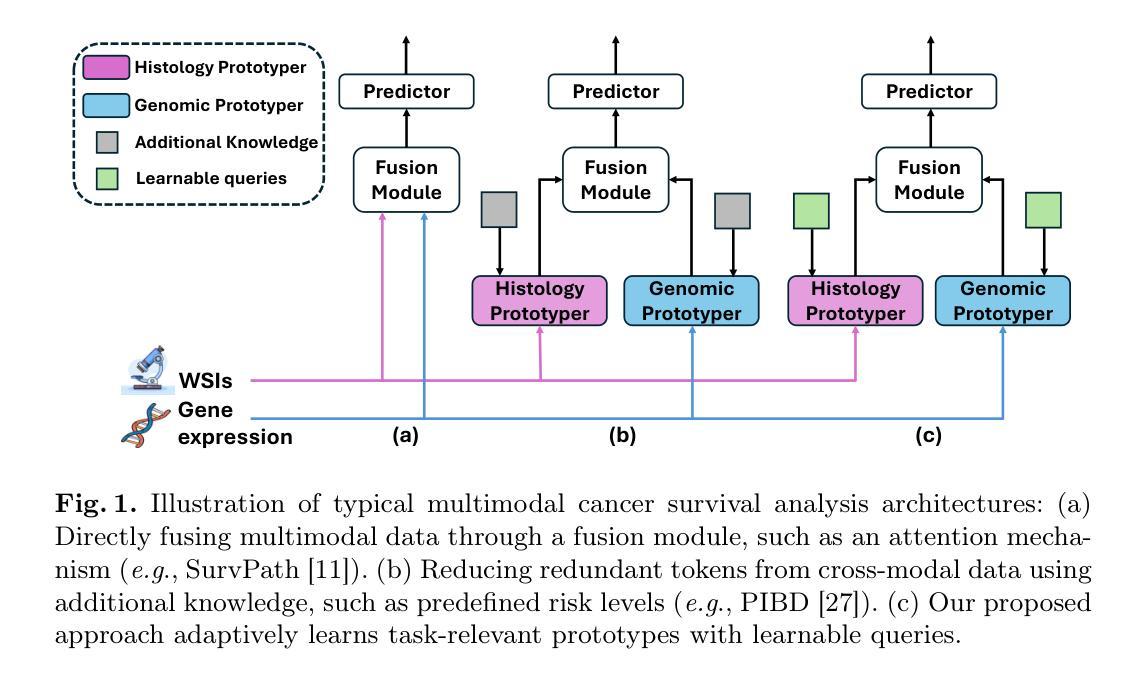
Adaptive Prototype Learning for Multimodal Cancer Survival Analysis
Authors:Hong Liu, Haosen Yang, Federica Eduati, Josien P. W. Pluim, Mitko Veta
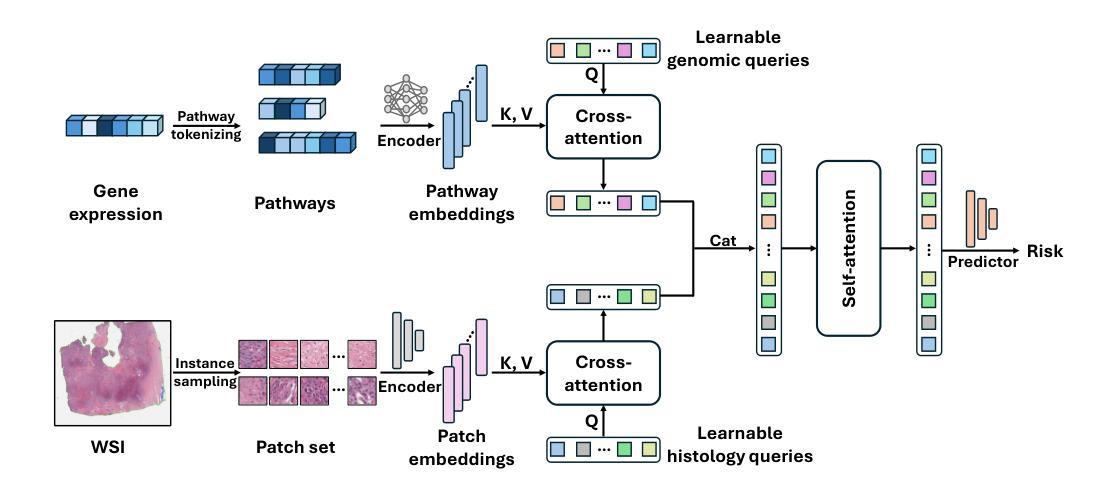
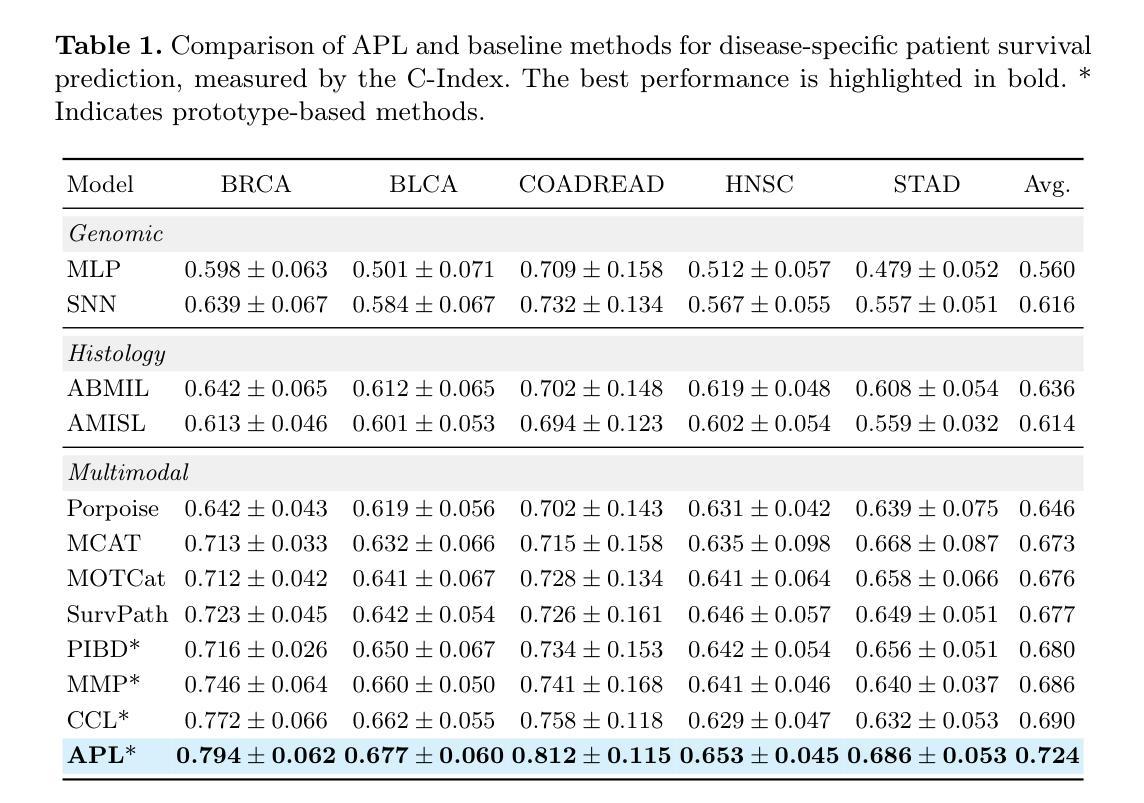
Leveraging multimodal data, particularly the integration of whole-slide histology images (WSIs) and transcriptomic profiles, holds great promise for improving cancer survival prediction. However, excessive redundancy in multimodal data can degrade model performance. In this paper, we propose Adaptive Prototype Learning (APL), a novel and effective approach for multimodal cancer survival analysis. APL adaptively learns representative prototypes in a data-driven manner, reducing redundancy while preserving critical information. Our method employs two sets of learnable query vectors that serve as a bridge between high-dimensional representations and survival prediction, capturing task-relevant features. Additionally, we introduce a multimodal mixed self-attention mechanism to enable cross-modal interactions, further enhancing information fusion. Extensive experiments on five benchmark cancer datasets demonstrate the superiority of our approach over existing methods. The code is available at https://github.com/HongLiuuuuu/APL.
利用多模态数据,特别是整合全切片组织图像(WSIs)和转录组图谱,在癌症生存预测方面展现出巨大潜力。然而,多模态数据中的过度冗余可能会降低模型性能。在本文中,我们提出了自适应原型学习(APL),这是一种用于多模态癌症生存分析的新型有效方法。APL以数据驱动的方式自适应地学习代表性原型,减少冗余的同时保留关键信息。我们的方法采用两组可学习的查询向量,作为高维表示和生存预测之间的桥梁,捕捉任务相关特征。此外,我们引入了一种多模态混合自注意机制,以实现跨模态交互,进一步增强信息融合。在五个基准癌症数据集上的广泛实验表明,我们的方法优于现有方法。代码可在https://github.com/HongLiuuuuu/APL获得。
论文及项目相关链接
PDF 10 pages, 3 figures
Summary
医学图像领域研究人员通过利用多模态数据,特别是整合全切片组织图像(WSIs)和转录组图谱,为改善癌症生存预测带来了希望。本文提出了一种自适应原型学习(APL)的新方法,能够有效处理多模态癌症生存分析中的数据冗余问题,自适应地学习代表性原型并保留关键信息。此外,引入了跨模态交互的自注意机制增强信息融合能力。代码已在GitHub上发布。该方法已在五个癌症数据集上进行实验验证。通过多维比较证明了该方法具有优异的预测效能和通用性。采用多任务联合学习增强特征选择能力的后续工作值得关注。不同医学分支疾病诊疗体系的相似性和差异需综合考虑,以实现更好的疾病诊疗融合研究。对于融合医学中的不同专业方向研究方法和策略也有待进一步探索和改进。未来的研究需要进一步加强不同疾病领域之间的交流和合作,以促进医学的全面发展。基于融合医学理念的交叉学科研究,对疾病的综合诊疗方案有着巨大潜力。在此背景下,借助大数据与人工智能的加持推进相关研究更具重要意义。本文主要讨论了癌症生存预测技术的现状和未来发展前景以及所面临的挑战,介绍了自己的创新性解决方案以及实践中的优点和挑战性特点。Key Takeaways:
- 利用多模态数据(全切片组织图像和转录组图谱)进行癌症生存预测具有巨大潜力。
- 自适应原型学习(APL)方法能有效处理多模态数据中的冗余信息,提高模型性能。
- APL方法引入跨模态交互的自注意机制以增强信息融合能力。
- 实验验证显示APL方法在五个癌症数据集上具有优异性能。
- 未来研究方向包括多任务联合学习增强特征选择能力、不同医学分支的疾病诊疗体系融合研究等。
- 融合医学理念下的交叉学科研究对疾病综合诊疗方案具有巨大潜力。大数据和人工智能技术在该领域的应用受到重视。当前工作主要讨论癌症生存预测技术的挑战和未来发展趋势。
点此查看论文截图

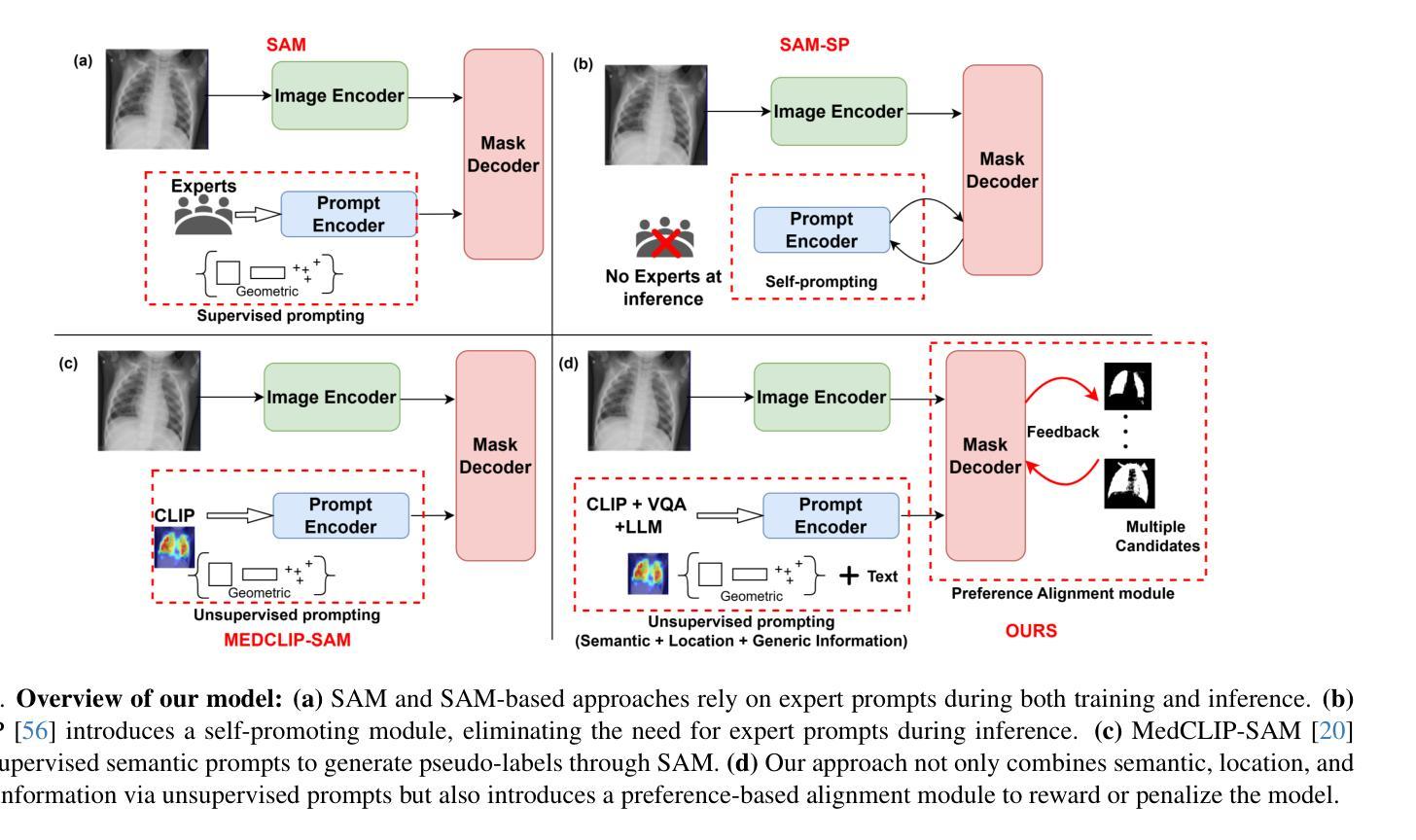
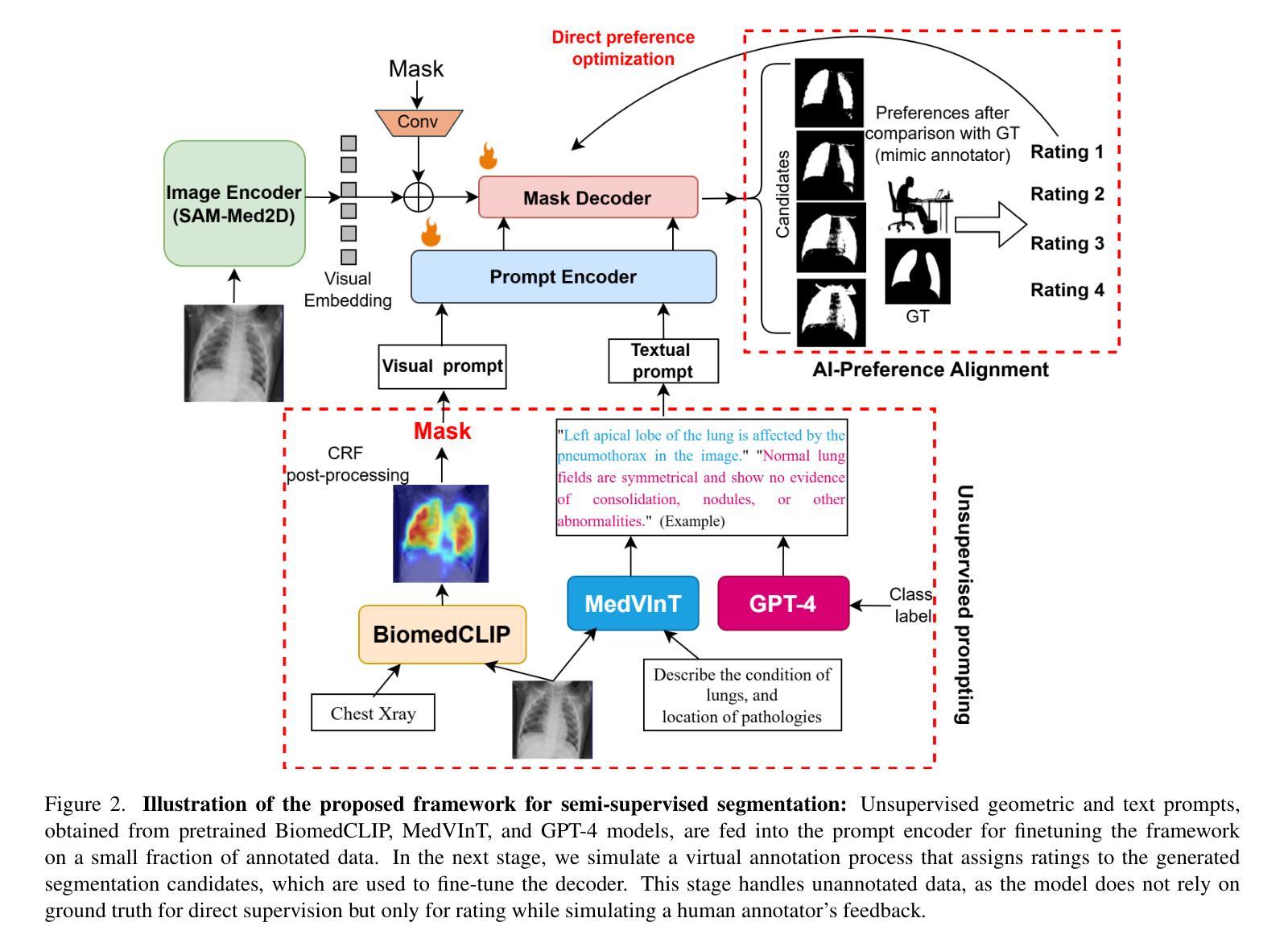
Enhancing SAM with Efficient Prompting and Preference Optimization for Semi-supervised Medical Image Segmentation
Authors:Aishik Konwer, Zhijian Yang, Erhan Bas, Cao Xiao, Prateek Prasanna, Parminder Bhatia, Taha Kass-Hout
Foundational models such as the Segment Anything Model (SAM) are gaining traction in medical imaging segmentation, supporting multiple downstream tasks. However, such models are supervised in nature, still relying on large annotated datasets or prompts supplied by experts. Conventional techniques such as active learning to alleviate such limitations are limited in scope and still necessitate continuous human involvement and complex domain knowledge for label refinement or establishing reward ground truth. To address these challenges, we propose an enhanced Segment Anything Model (SAM) framework that utilizes annotation-efficient prompts generated in a fully unsupervised fashion, while still capturing essential semantic, location, and shape information through contrastive language-image pretraining and visual question answering. We adopt the direct preference optimization technique to design an optimal policy that enables the model to generate high-fidelity segmentations with simple ratings or rankings provided by a virtual annotator simulating the human annotation process. State-of-the-art performance of our framework in tasks such as lung segmentation, breast tumor segmentation, and organ segmentation across various modalities, including X-ray, ultrasound, and abdominal CT, justifies its effectiveness in low-annotation data scenarios.
像Segment Anything Model(SAM)这样的基础模型在医学图像分割中越来越受欢迎,支持多个下游任务。然而,此类模型本质上是监督的,仍然依赖于专家提供的大量标注数据集或提示。为了缓解这些限制,采用主动学习的传统技术范围有限,仍需要持续的人力投入和复杂的领域知识来进行标签细化或建立奖励基准真实值。为了解决这些挑战,我们提出了一种增强的Segment Anything Model(SAM)框架,该框架利用完全无监督生成的标注有效提示,同时通过对比语言图像预训练和视觉问答捕获关键的语义、位置和形状信息。我们采用直接偏好优化技术来设计最佳策略,使模型能够借助模拟人类注释过程的虚拟注释器提供的简单评分或排名生成高保真分割。我们的框架在肺分割、乳腺肿瘤分割和器官分割等任务中的最新性能表现,以及在X射线、超声和腹部CT等各种模态的应用,证明了它在低标注数据场景中的有效性。
论文及项目相关链接
PDF Accepted to CVPR 2025
Summary
基于Segment Anything Model(SAM)的基础模型在医学图像分割中逐渐受到重视,支持多任务处理。然而,此类模型仍依赖于大量标注数据集或专家提供的提示,存在监督学习的局限性。为解决这些问题,我们提出了一种增强的SAM框架,该框架以完全无监督的方式生成标注有效的提示,同时仍能通过对比语言图像预训练和视觉问答捕获关键的语义、位置和形状信息。采用直接偏好优化技术设计最佳策略,使模型能够借助模拟人类注释过程的虚拟注释器提供的简单评分或排名生成高保真分割。在肺分割、乳腺肿瘤分割和器官分割等任务中,该框架的先进性能证明了其在低标注数据场景中的有效性。
Key Takeaways
- Segment Anything Model (SAM) 在医学图像分割中受到重视,支持多任务处理。
- SAM仍依赖于大量标注数据和专家提示,存在监督学习的局限性。
- 提出的增强SAM框架以完全无监督的方式生成标注有效的提示。
- 该框架通过对比语言图像预训练和视觉问答捕获关键的语义、位置和形状信息。
- 采用直接偏好优化技术设计最佳策略,生成高保真分割。
- 框架在肺分割、乳腺肿瘤分割和器官分割等任务中表现出先进性能。
- 框架在低标注数据场景中的有效性得到证明。
点此查看论文截图

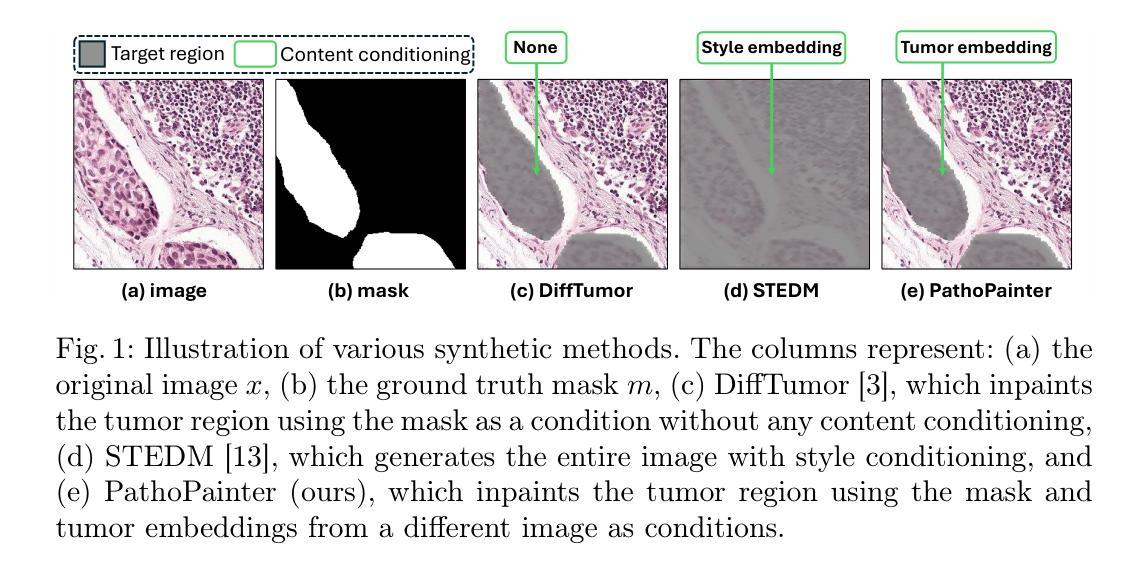
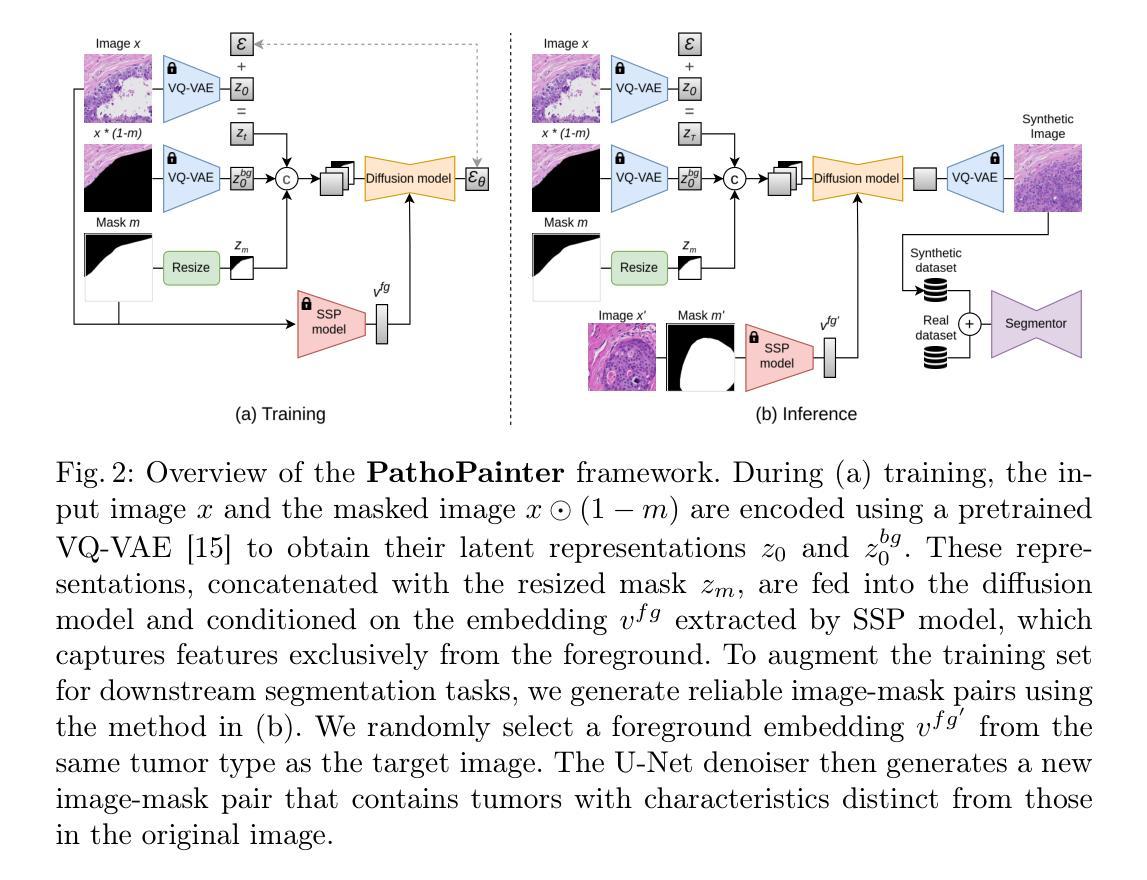
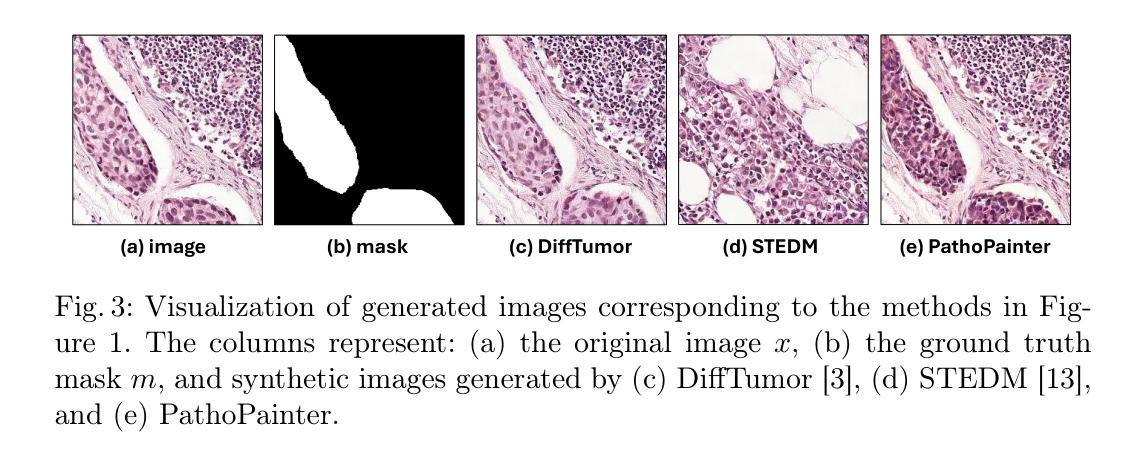
PathoPainter: Augmenting Histopathology Segmentation via Tumor-aware Inpainting
Authors:Hong Liu, Haosen Yang, Evi M. C. Huijben, Mark Schuiveling, Ruisheng Su, Josien P. W. Pluim, Mitko Veta
Tumor segmentation plays a critical role in histopathology, but it requires costly, fine-grained image-mask pairs annotated by pathologists. Thus, synthesizing histopathology data to expand the dataset is highly desirable. Previous works suffer from inaccuracies and limited diversity in image-mask pairs, both of which affect training segmentation, particularly in small-scale datasets and the inherently complex nature of histopathology images. To address this challenge, we propose PathoPainter, which reformulates image-mask pair generation as a tumor inpainting task. Specifically, our approach preserves the background while inpainting the tumor region, ensuring precise alignment between the generated image and its corresponding mask. To enhance dataset diversity while maintaining biological plausibility, we incorporate a sampling mechanism that conditions tumor inpainting on regional embeddings from a different image. Additionally, we introduce a filtering strategy to exclude uncertain synthetic regions, further improving the quality of the generated data. Our comprehensive evaluation spans multiple datasets featuring diverse tumor types and various training data scales. As a result, segmentation improved significantly with our synthetic data, surpassing existing segmentation data synthesis approaches, e.g., 75.69% -> 77.69% on CAMELYON16. The code is available at https://github.com/HongLiuuuuu/PathoPainter.
肿瘤分割在病理学中扮演着至关重要的角色,但它需要病理学家标注的昂贵、精细的图像-掩膜对。因此,合成病理数据以扩大数据集是非常理想的。之前的工作存在图像-掩膜对的不准确性和多样性有限的问题,这两者都会影响分割训练,特别是在小规模数据集和病理图像固有的复杂性方面。为了应对这一挑战,我们提出了PathoPainter,它将图像-掩膜对的生成重新制定为一个肿瘤修复任务。具体来说,我们的方法在修复肿瘤区域的同时保留背景,确保生成的图像与其对应的掩膜之间精确对齐。为了在保持生物合理性的同时提高数据集的多样性,我们引入了一种采样机制,该机制根据来自不同图像的局部嵌入来进行肿瘤修复。此外,我们还采用了一种过滤策略,以排除不确定的合成区域,进一步提高生成数据的质量。我们的综合评估涵盖了多个包含各种肿瘤类型和不同训练数据规模的数据集。因此,使用我们的合成数据,分割效果得到了显著提高,超越了现有的分割数据合成方法,例如在CAMELYON16上的成绩从75.69%提高到了77.69%。代码可通过https://github.com/HongLiuuuuu/PathoPainter获取。
论文及项目相关链接
PDF 10 pages, 3 figures
Summary
本文提出一种基于肿瘤修复技术(PathoPainter)的数据集合成方法,旨在解决肿瘤分割领域中对病理学专家精细标注的图像-掩膜对数据的依赖问题。该方法通过肿瘤区域修复技术生成图像和掩膜,确保生成的图像与掩膜精确对齐,同时引入采样机制增加数据集多样性并保持生物学合理性。通过过滤不确定的合成分割区域,提高生成数据质量。在多个数据集上的评估显示,使用合成数据可显著提高分割性能,超越现有分割数据合成方法。
Key Takeaways
- PathoPainter是一种用于合成肿瘤分割数据集的方法,旨在解决数据获取成本高的问题。
- 通过肿瘤修复技术生成图像和掩膜,确保精确对齐。
- 采用采样机制增加数据集多样性并维持生物学合理性。
- 引入过滤策略以排除不确定的合成分割区域。
- 在多个数据集上的评估显示,使用PathoPainter合成的数据能显著提高分割性能。
- 该方法性能优于现有的分割数据合成方法。
点此查看论文截图

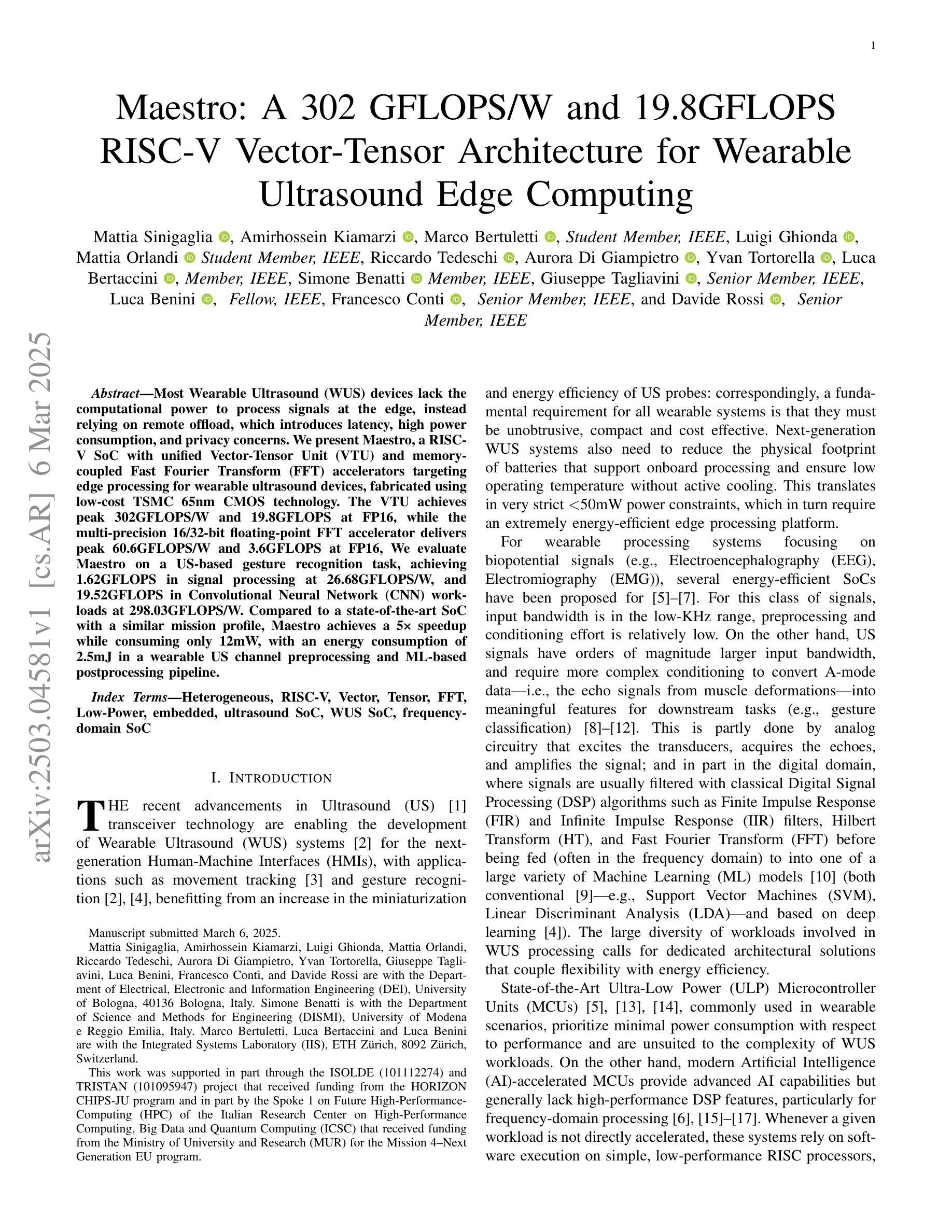
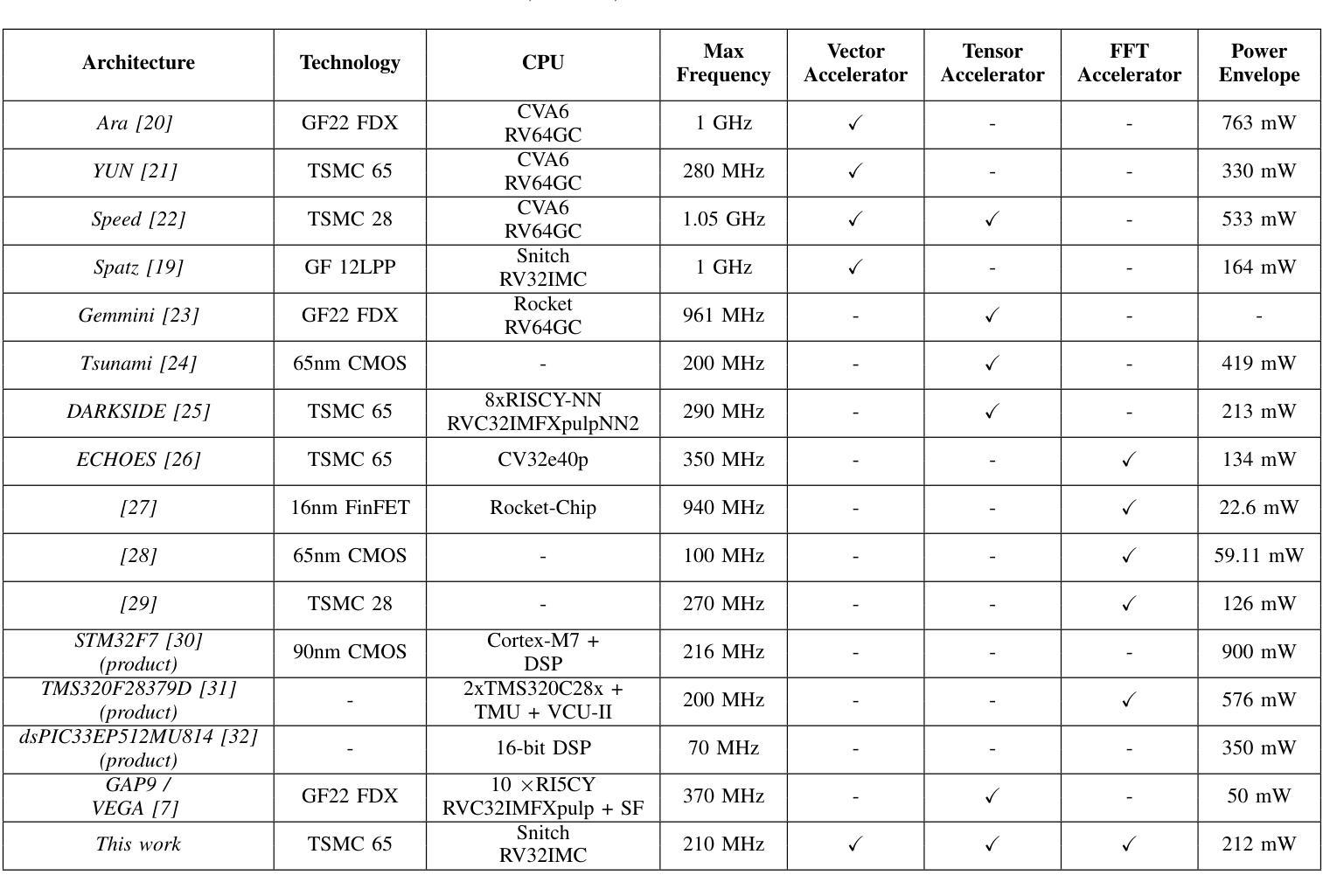
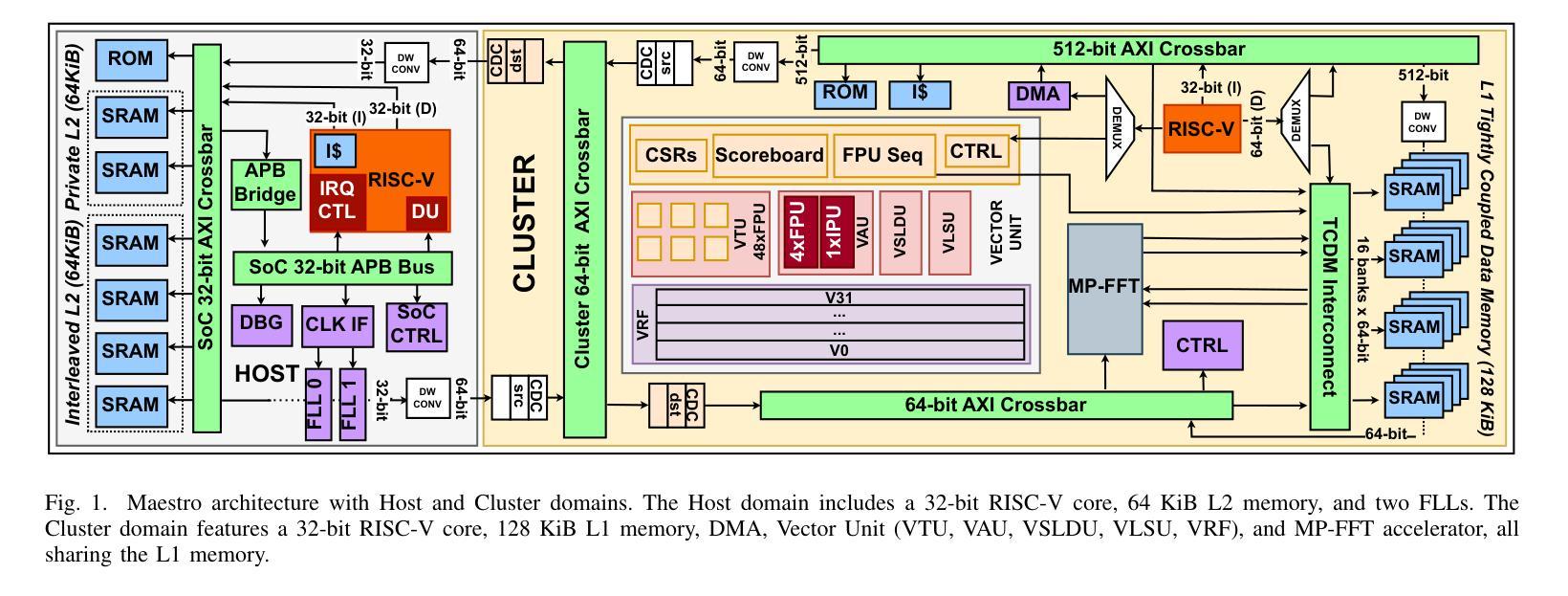
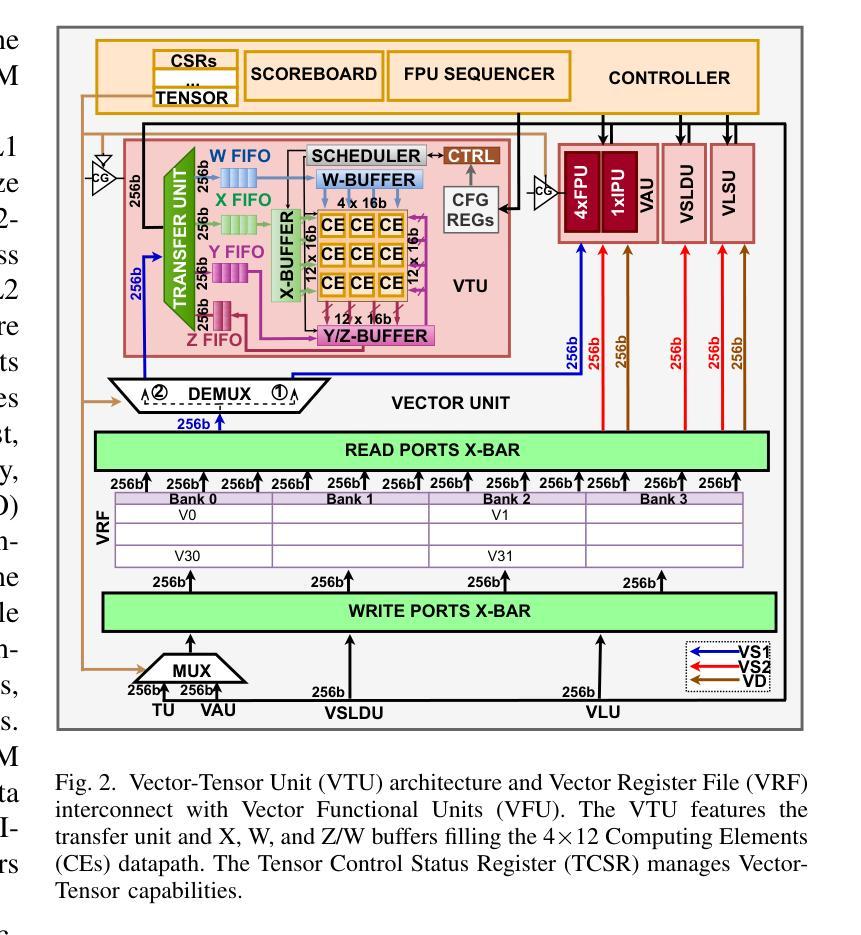
Maestro: A 302 GFLOPS/W and 19.8GFLOPS RISC-V Vector-Tensor Architecture for Wearable Ultrasound Edge Computing
Authors:Mattia Sinigaglia, Amirhossein Kiamarzi, Marco Bertuletti, Luigi Ghionda, Mattia Orlandi, Riccardo Tedeschi, Aurora Di Giampietro, Yvan Tortorella, Luca Bertaccini, Simone Benatti, Giuseppe Tagliavini, Luca Benini, Francesco Conti, Davide Rossi
Most Wearable Ultrasound (WUS) devices lack the computational power to process signals at the edge, instead relying on remote offload, which introduces latency, high power consumption, and privacy concerns. We present Maestro, a RISC-V SoC with unified Vector-Tensor Unit (VTU) and memory-coupled Fast Fourier Transform (FFT) accelerators targeting edge processing for wearable ultrasound devices, fabricated using low-cost TSMC 65nm CMOS technology. The VTU achieves peak 302GFLOPS/W and 19.8GFLOPS at FP16, while the multi-precision 16/32-bit floating-point FFT accelerator delivers peak 60.6GFLOPS/W and 3.6GFLOPS at FP16, We evaluate Maestro on a US-based gesture recognition task, achieving 1.62GFLOPS in signal processing at 26.68GFLOPS/W, and 19.52GFLOPS in Convolutional Neural Network (CNN) workloads at 298.03GFLOPS/W. Compared to a state-of-the-art SoC with a similar mission profile, Maestro achieves a 5x speedup while consuming only 12mW, with an energy consumption of 2.5mJ in a wearable US channel preprocessing and ML-based postprocessing pipeline.
大多数可穿戴超声(WUS)设备缺乏在边缘处理信号的计算能力,而是依赖于远程卸载,这引入了延迟、高功耗和隐私担忧。我们推出了Maestro,这是一款针对可穿戴超声设备的边缘处理而设计的RISC-V系统芯片(SoC),配备了统一的向量张量单元(VTU)和内存耦合快速傅里叶变换(FFT)加速器,采用低成本TSMC 65nm CMOS技术制造。VTU在FP16下达到峰值302GFLOPS/W和19.8GFLOPS,而多精度16/32位浮点FFT加速器在FP16下达到峰值60.6GFLOPS/W和3.6GFLOPS。我们评估了Maestro在基于美国的姿态识别任务上的表现,在信号处理的GFLOPS达到26.68时实现了1.62GFLOPS的处理速度,在卷积神经网络(CNN)工作负载的GFLOPS达到298.03时实现了19.52GFLOPS的处理速度。与具有类似任务配置的最新SoC相比,Maestro实现了5倍的加速,仅消耗12mW的功耗,在可穿戴超声通道预处理和基于机器学习的后处理管道中的能耗为2.5mJ。
论文及项目相关链接
Summary
一种新型的穿戴式超声处理系统Maestro被研发出来,它具备边缘处理能力,解决了现有穿戴式超声设备依赖远程处理导致的延迟、高功耗和隐私等问题。Maestro采用RISC-V SoC,配备了统一的向量张量单元(VTU)和内存耦合快速傅里叶变换(FFT)加速器,实现了低功耗处理。其在手势识别任务中表现优异,相比现有最先进的类似任务SoC,Maestro实现了5倍的加速,同时功耗仅为12mW。
Key Takeaways
- Maestro是一种新型的穿戴式超声处理系统,具备边缘处理能力。
- Maestro解决了现有穿戴式超声设备依赖远程处理的问题,减少了延迟、高功耗和隐私担忧。
- Maestro采用RISC-V SoC,配备了VTU和FFT加速器,实现了低功耗处理。
- VTU实现了峰值302GFLOPS/W和19.8GFLOPS的计算能力。
- FFT加速器提供了多精度计算,峰值达到60.6GFLOPS/W。
- 在手势识别任务中,Maestro相比现有最先进的SoC实现了5倍的加速。
点此查看论文截图
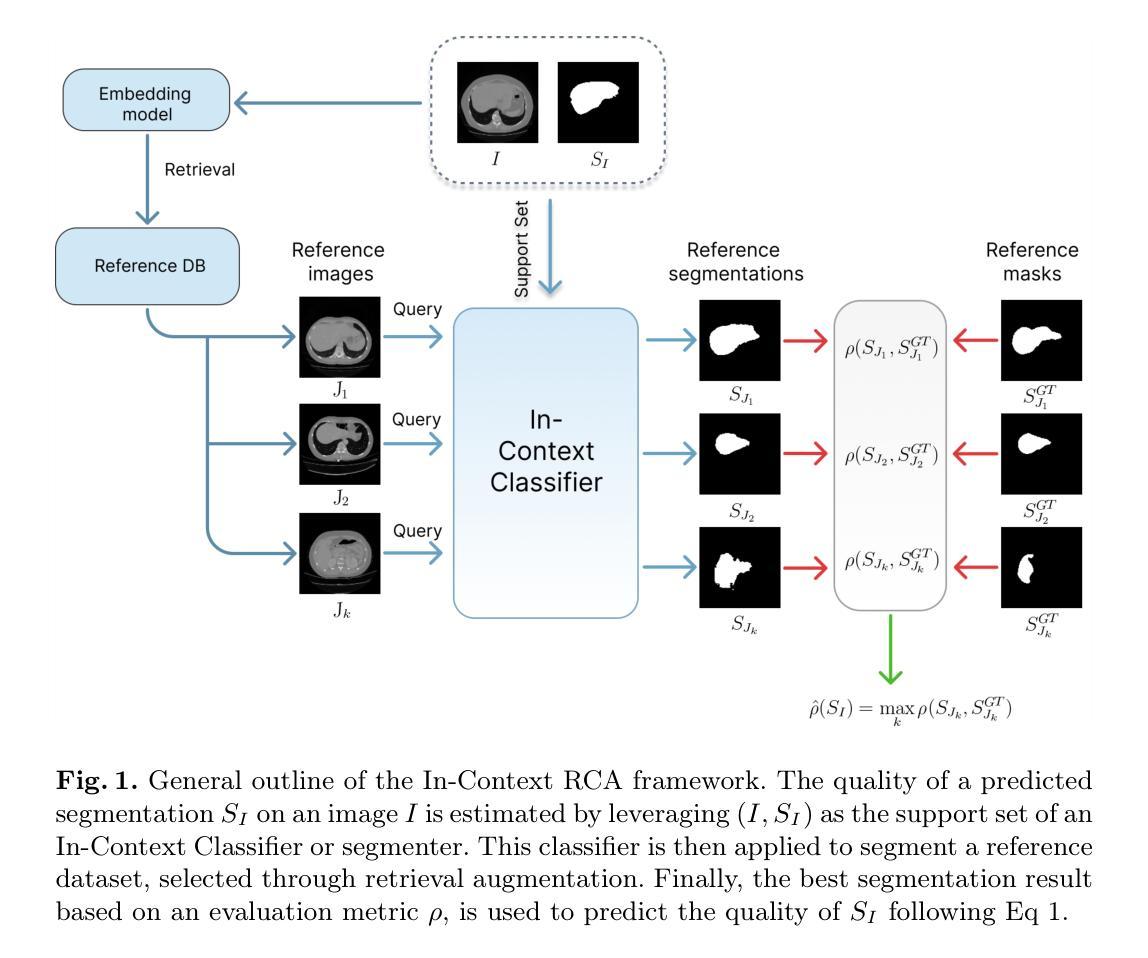
In-Context Reverse Classification Accuracy: Efficient Estimation of Segmentation Quality without Ground-Truth
Authors:Matias Cosarinsky, Ramiro Billot, Lucas Mansilla, Gabriel Gimenez, Nicolas Gaggión, Guanghui Fu, Enzo Ferrante
Assessing the quality of automatic image segmentation is crucial in clinical practice, but often very challenging due to the limited availability of ground truth annotations. In this paper, we introduce In-Context Reverse Classification Accuracy (In-Context RCA), a novel framework for automatically estimating segmentation quality in the absence of ground-truth annotations. By leveraging recent in-context learning segmentation models and incorporating retrieval-augmentation techniques to select the most relevant reference images, our approach enables efficient quality estimation with minimal reference data. Validated across diverse medical imaging modalities, our method demonstrates robust performance and computational efficiency, offering a promising solution for automated quality control in clinical workflows, where fast and reliable segmentation assessment is essential. The code is available at https://github.com/mcosarinsky/In-Context-RCA.
在医疗实践中,评估自动图像分割的质量至关重要,但由于真实标注数据的有限性,这通常极具挑战性。在本文中,我们引入了基于上下文反向分类准确率(In-Context RCA)这一新型框架,无需真实标注数据即可自动估计分割质量。通过利用最新的上下文学习分割模型和检索增强技术来选择最相关的参考图像,我们的方法能够在最少的参考数据下实现高效的质量评估。经过多种医学影像模态的验证,我们的方法表现出了稳健的性能和计算效率,为临床工作流程中的自动化质量控制提供了有前景的解决方案,特别是在快速可靠的分割评估至关重要的领域。相关代码可访问https://github.com/mcosarinsky/In-Context-RCA。
论文及项目相关链接
Summary
医学图像自动分割质量评估在临床实践中至关重要,但缺乏真实标注导致评估具有挑战性。本文提出一种新型框架In-Context Reverse Classification Accuracy(In-Context RCA),无需真实标注即可自动估计分割质量。该框架结合最新的上下文学习分割模型和检索增强技术,选择最相关的参考图像,以最小的参考数据实现高效的质量评估。经过多种医学成像模态的验证,该方法表现出稳健的性能和计算效率,为临床工作流程中的自动化质量控制提供了有前景的解决方案,尤其在需要快速可靠分割评估的情况下。
Key Takeaways
- 医学图像自动分割质量评估的重要性及挑战。
- 提出了一种新型框架In-Context RCA,无需真实标注即可评估分割质量。
- 利用最新的上下文学习分割模型。
- 引入检索增强技术,选择最相关的参考图像。
- 方法在多种医学成像模态下表现出稳健性和计算效率。
- 对临床工作流程的自动化质量控制具有潜在的应用价值。
点此查看论文截图

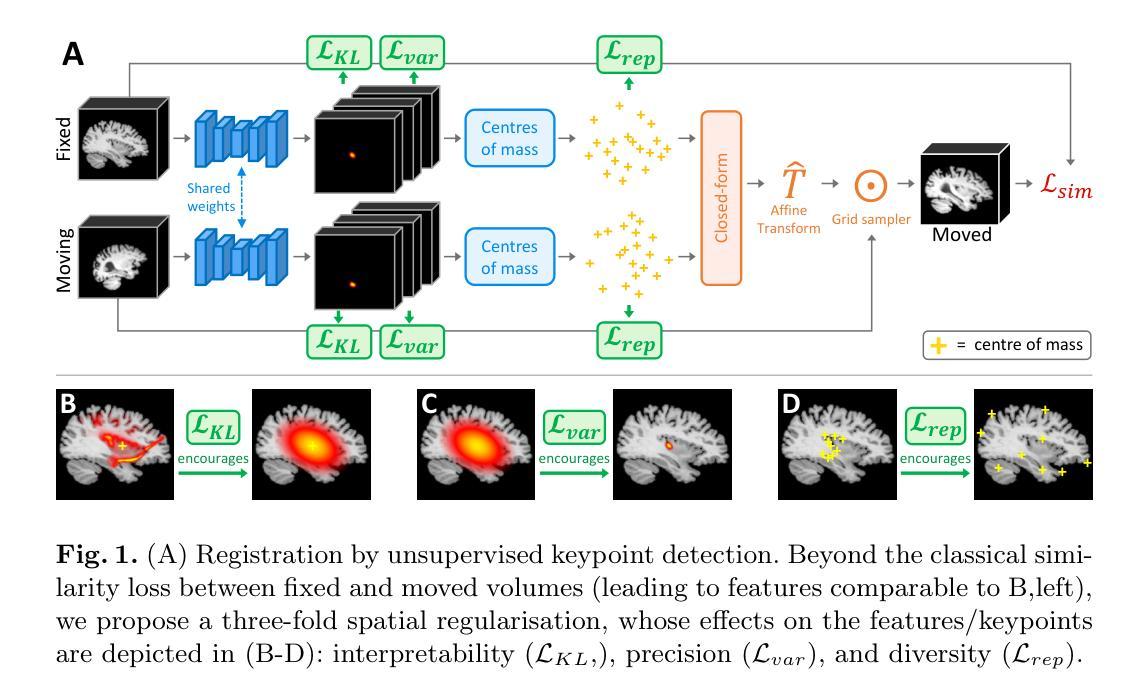
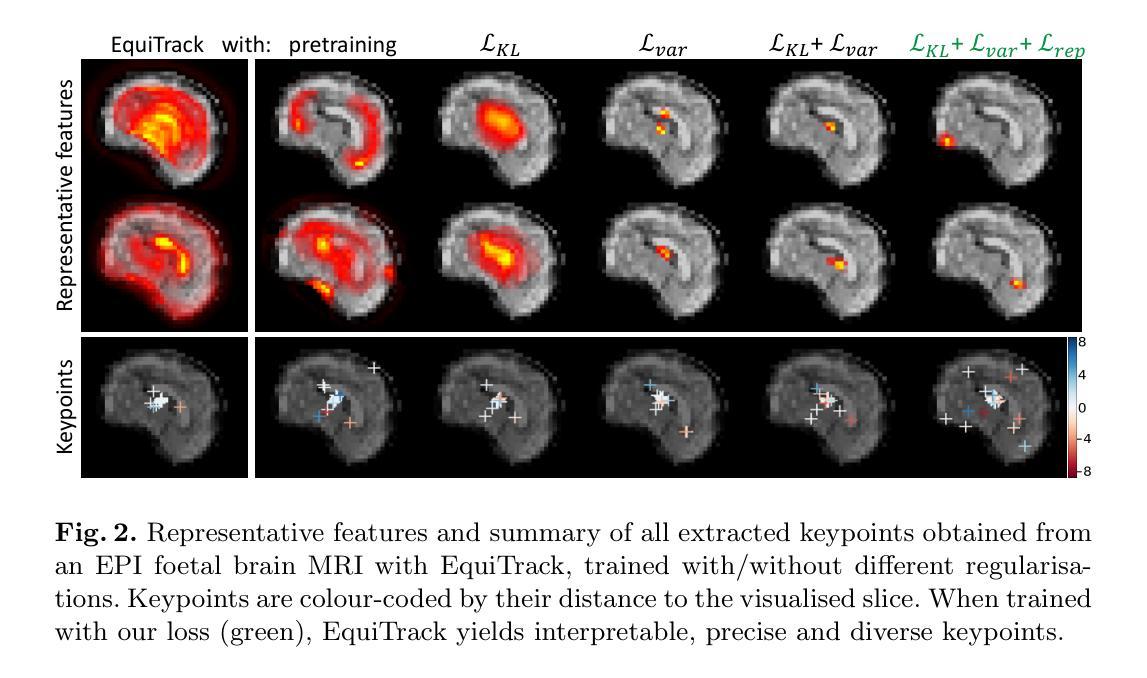
Spatial regularisation for improved accuracy and interpretability in keypoint-based registration
Authors:Benjamin Billot, Ramya Muthukrishnan, Esra Abaci-Turk, Ellen P. Grant, Nicholas Ayache, Hervé Delingette, Polina Golland
Unsupervised registration strategies bypass requirements in ground truth transforms or segmentations by optimising similarity metrics between fixed and moved volumes. Among these methods, a recent subclass of approaches based on unsupervised keypoint detection stand out as very promising for interpretability. Specifically, these methods train a network to predict feature maps for fixed and moving images, from which explainable centres of mass are computed to obtain point clouds, that are then aligned in closed-form. However, the features returned by the network often yield spatially diffuse patterns that are hard to interpret, thus undermining the purpose of keypoint-based registration. Here, we propose a three-fold loss to regularise the spatial distribution of the features. First, we use the KL divergence to model features as point spread functions that we interpret as probabilistic keypoints. Then, we sharpen the spatial distributions of these features to increase the precision of the detected landmarks. Finally, we introduce a new repulsive loss across keypoints to encourage spatial diversity. Overall, our loss considerably improves the interpretability of the features, which now correspond to precise and anatomically meaningful landmarks. We demonstrate our three-fold loss in foetal rigid motion tracking and brain MRI affine registration tasks, where it not only outperforms state-of-the-art unsupervised strategies, but also bridges the gap with state-of-the-art supervised methods. Our code is available at https://github.com/BenBillot/spatial_regularisation.
无监督的注册策略通过优化固定体积和移动体积之间的相似性度量来绕过对真实变换或分割的要求。在这些方法中,最近出现的一种基于无监督关键点检测的方法作为解释性方面非常有前途的一个子类而脱颖而出。具体来说,这些方法训练网络为固定图像和移动图像预测特征图,从中计算出可解释的质量中心,以获得点云,然后这些点云以封闭形式对齐。然而,网络返回的特征通常会产生难以解释的空间扩散模式,从而破坏了基于关键点的注册目的。在这里,我们提出了一个三重的损失来正规化特征的空间分布。首先,我们使用KL散度来模拟特征作为点扩散函数,我们将其解释为概率性的关键点。然后,我们使这些特征的空间分布更加清晰,以提高检测到的地标的精度。最后,我们在关键点之间引入了一个新的排斥损失来鼓励空间多样性。总的来说,我们的损失大大提高了特征的可解释性,这些特征现在对应于精确且解剖上有意义的地标。我们在胎儿刚性运动跟踪和脑MRI仿射注册任务中展示了我们的三重损失,它不仅优于最新的无监督策略,而且缩小了与最新监督方法的差距。我们的代码可在https://github.com/BenBillot/spatial_regularisation 中找到。
论文及项目相关链接
PDF under review
摘要
本文提出一种基于无监督关键点检测的方法,通过优化固定图像和移动图像之间的相似性度量来实现图像配准,同时提出了一种新的三重损失函数对特征的空间分布进行正则化,提高了解释性关键点检测的准确性。实验结果表明,该方法不仅优于现有的无监督策略,而且缩小了与最新监督方法的差距。代码已公开。
关键发现点
- 提出了一种基于无监督关键点检测的无监督配准方法。
- 通过训练网络预测固定和移动图像的特征映射,并计算可解释的中心质量来获取点云进行对齐。
- 通过KL散度将特征建模为点扩散函数,解释为概率关键点。
- 对特征的空间分布进行锐化以提高检测到的地标精度。
- 引入了一种新的排斥损失来鼓励关键点的空间多样性。
- 三重损失函数显著提高了特征的解释性,这些特征现在对应于精确且解剖上意义的地标。
点此查看论文截图

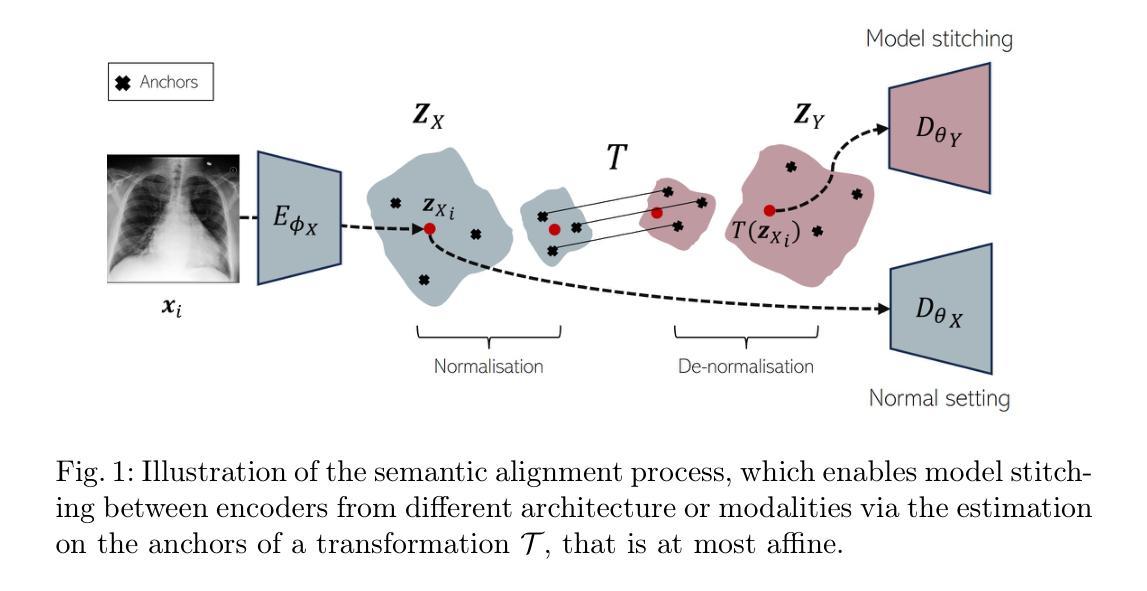
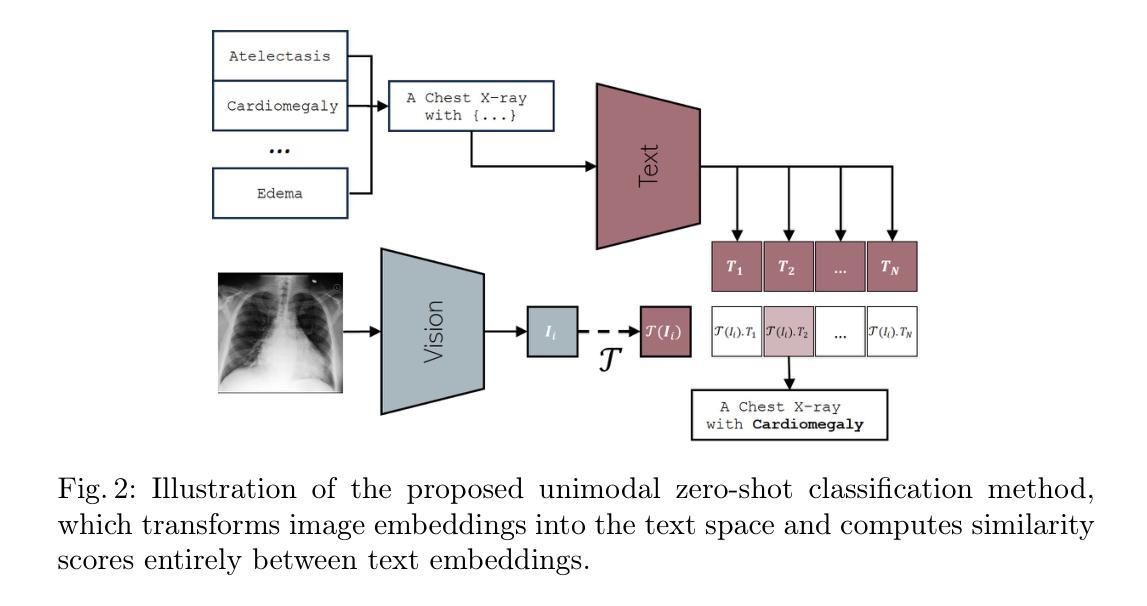
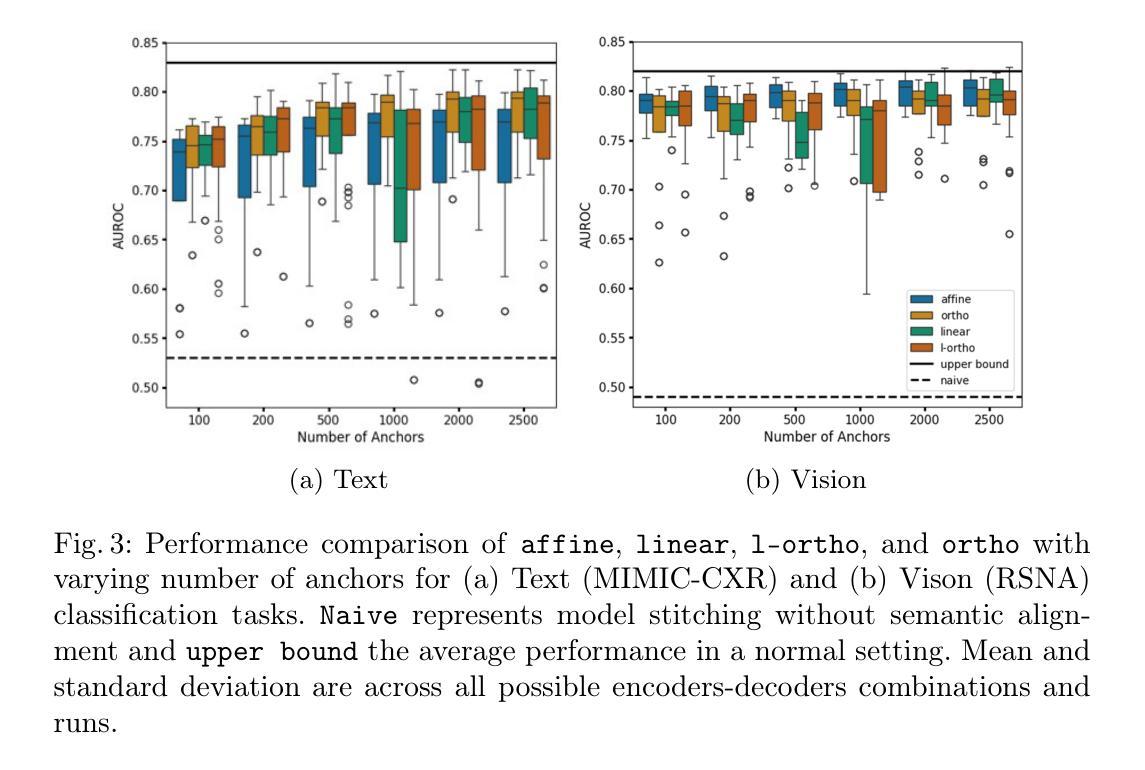
Semantic Alignment of Unimodal Medical Text and Vision Representations
Authors:Maxime Di Folco, Emily Chan, Marta Hasny, Cosmin I. Bercea, Julia A. Schnabel
General-purpose AI models, particularly those designed for text and vision, demonstrate impressive versatility across a wide range of deep-learning tasks. However, they often underperform in specialised domains like medical imaging, where domain-specific solutions or alternative knowledge transfer approaches are typically required. Recent studies have noted that general-purpose models can exhibit similar latent spaces when processing semantically related data, although this alignment does not occur naturally. Building on this insight, it has been shown that applying a simple transformation - at most affine - estimated from a subset of semantically corresponding samples, known as anchors, enables model stitching across diverse training paradigms, architectures, and modalities. In this paper, we explore how semantic alignment - estimating transformations between anchors - can bridge general-purpose AI with specialised medical knowledge. Using multiple public chest X-ray datasets, we demonstrate that model stitching across model architectures allows general models to integrate domain-specific knowledge without additional training, leading to improved performance on medical tasks. Furthermore, we introduce a novel zero-shot classification approach for unimodal vision encoders that leverages semantic alignment across modalities. Our results show that our method not only outperforms general multimodal models but also approaches the performance levels of fully trained, medical-specific multimodal solutions
通用人工智能模型,尤其是那些为文本和视觉设计的模型,在广泛的深度学习任务中表现出了令人印象深刻的通用性。然而,它们在专业领域如医学影像中常常表现不佳,这通常需要领域特定的解决方案或替代知识迁移方法。近期的研究发现,当处理语义相关数据时,通用模型可以展现出类似的潜在空间,尽管这种对齐并不会自然发生。基于这一见解,已经证明应用一种简单的转换——最多是仿射变换——通过从语义对应的样本子集估计出来,这些样本被称为锚点,可以在不同的训练模式、架构和模态之间进行模型拼接。在本文中,我们探讨了语义对齐如何估算锚点之间的转换——能够架起通用人工智能与特殊医学知识之间的桥梁。通过使用多个公共胸部X射线数据集,我们证明了跨模型架构的模型拼接允许通用模型整合特定领域的知识而无需额外训练,从而提高了医学任务上的性能。此外,我们介绍了一种新型的零样本分类方法,用于单模态视觉编码器,该方法利用跨模态的语义对齐。我们的结果表明,我们的方法不仅优于一般的跨模态模型,而且接近经过充分训练的特定医学跨模态解决方案的性能水平。
论文及项目相关链接
Summary
本文探讨了通用人工智能模型在医学成像等特定领域的应用挑战。通过语义对齐技术,实现在不同训练范式、架构和模态之间的模型拼接,使通用模型能够整合特定领域知识,提高在医疗任务上的性能。同时,提出了一种基于语义对齐的零射击分类方法,对于单模态视觉编码器性能有明显提升,接近全训练的医学专用多模态解决方案。
Key Takeaways
- 通用人工智能模型在医学成像等特定领域存在性能瓶颈,需要特定领域的解决方案或知识迁移方法。
- 语义对齐技术能够实现模型拼接,使得通用模型整合特定领域知识成为可能。
- 通过使用锚点进行语义对齐估计,可以实现简单的模型转换,提高模型性能。
- 在多个公共胸部X光数据集上,模型架构之间的拼接使得通用模型能够融入特定领域知识,无需额外训练。
- 提出了一种新型的零射击分类方法,利用跨模态的语义对齐,对单模态视觉编码器的性能有显著提升。
- 该方法不仅优于一般的多模态模型,而且接近完全训练的医学专用多模态解决方案的性能水平。
点此查看论文截图

GBT-SAM: A Parameter-Efficient Depth-Aware Model for Generalizable Brain tumour Segmentation on mp-MRI
Authors:Cecilia Diana-Albelda, Roberto Alcover-Couso, Álvaro García-Martín, Jesus Bescos, Marcos Escudero-Viñolo
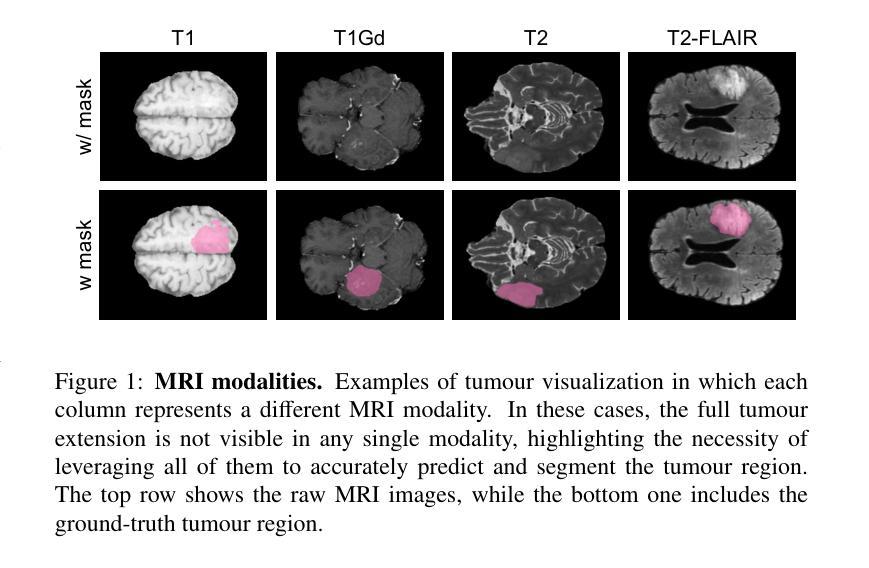
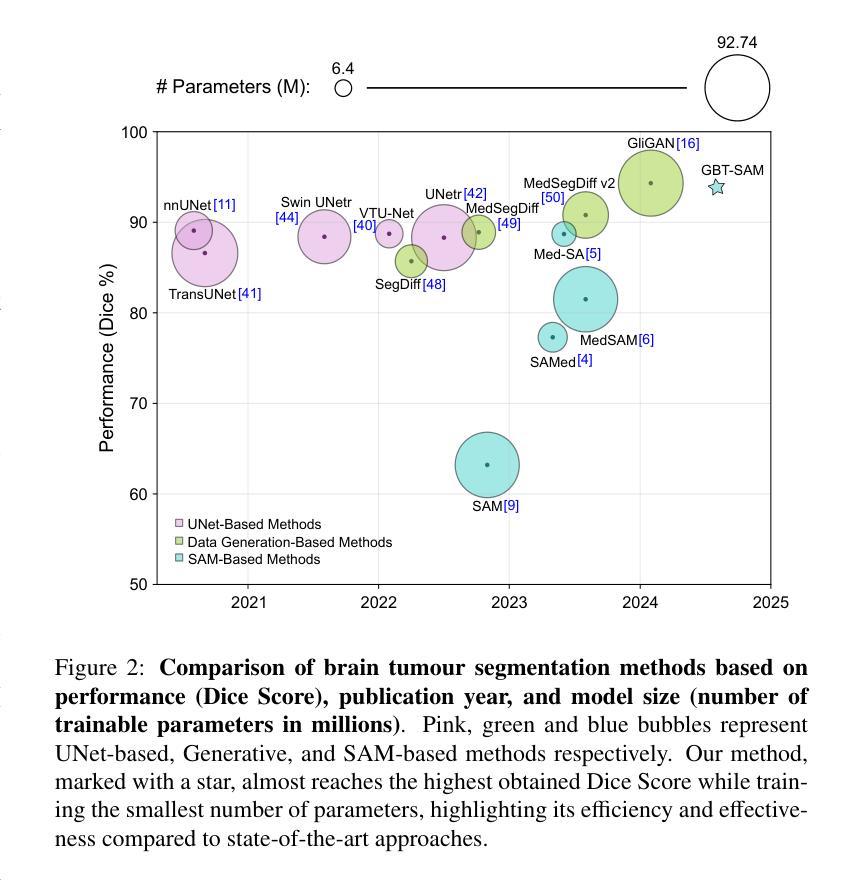
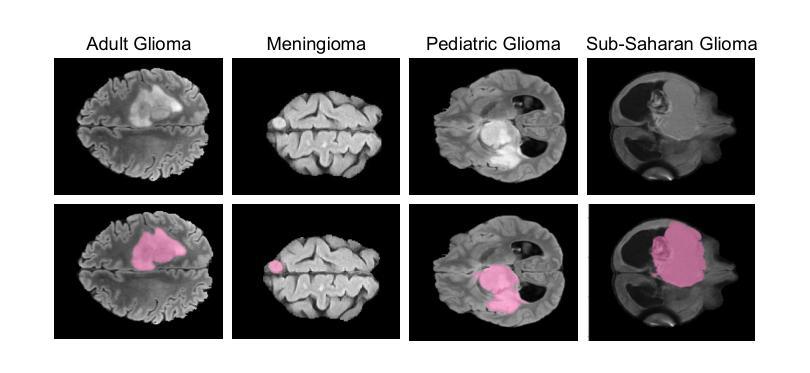
Gliomas are brain tumours that stand out for their highly lethal and aggressive nature, which demands a precise approach in their diagnosis. Medical image segmentation plays a crucial role in the evaluation and follow-up of these tumours, allowing specialists to analyse their morphology. However, existing methods for automatic glioma segmentation often lack generalization capability across other brain tumour domains, require extensive computational resources, or fail to fully utilize the multi-parametric MRI (mp-MRI) data used to delineate them. In this work, we introduce GBT-SAM, a novel Generalizable Brain Tumour (GBT) framework that extends the Segment Anything Model (SAM) to brain tumour segmentation tasks. Our method employs a two-step training protocol: first, fine-tuning the patch embedding layer to process the entire mp-MRI modalities, and second, incorporating parameter-efficient LoRA blocks and a Depth-Condition block into the Vision Transformer (ViT) to capture inter-slice correlations. GBT-SAM achieves state-of-the-art performance on the Adult Glioma dataset (Dice Score of $93.54$) while demonstrating robust generalization across Meningioma, Pediatric Glioma, and Sub-Saharan Glioma datasets. Furthermore, GBT-SAM uses less than 6.5M trainable parameters, thus offering an efficient solution for brain tumour segmentation. \ Our code and models are available at https://github.com/vpulab/med-sam-brain .
胶质瘤是突出的脑肿瘤,具有高度的致死性和侵袭性,这就要求在诊断时需要采取精确的方法。医学图像分割在评估和随访这些肿瘤中起着至关重要的作用,允许专家分析它们的形态。然而,现有的自动胶质瘤分割方法往往缺乏在其他脑肿瘤领域的泛化能力,需要大量的计算资源,或者未能充分利用用于界定肿瘤的多参数MRI(mp-MRI)数据。在这项工作中,我们介绍了GBT-SAM,这是一个可泛化脑肿瘤(GBT)的新框架,它扩展了分段任何模型(SAM)以用于脑肿瘤分割任务。我们的方法采用两步训练协议:首先,微调补丁嵌入层以处理整个mp-MRI模式;其次,在视觉变压器(ViT)中融入参数高效的LoRA块和深度条件块,以捕捉切片间的相关性。GBT-SAM在成人胶质瘤数据集上实现了最先进的性能(Dice得分为93.54%),同时在脑膜瘤、儿童胶质瘤和撒哈拉以南胶质瘤数据集上表现出稳健的泛化能力。此外,GBT-SAM使用的可训练参数少于650万,因此为脑肿瘤分割提供了有效的解决方案。我们的代码和模型可在https://github.com/vpulab/med-sam-brain上找到。
论文及项目相关链接
Summary
本文介绍了一种名为GBT-SAM的新型通用脑肿瘤(GBT)分割框架,该框架扩展了Segment Anything Model(SAM)以应用于脑肿瘤分割任务。它采用了一种两步训练协议,通过在Patch嵌入层处理整个多参数MRI(mp-MRI)模式并进行精细调整,以及将参数高效的LoRA块和深度条件块融入视觉转换器(ViT)以捕获切片间的相关性。GBT-SAM在成人胶质瘤数据集上实现了最佳性能(Dice分数为93.54%),并在脑膜瘤、儿童胶质瘤和撒哈拉以南胶质瘤数据集上表现出稳健的泛化能力。此外,GBT-SAM使用的可训练参数少于650万,为脑肿瘤分割提供了高效的解决方案。
Key Takeaways
- GBT-SAM是一个针对脑肿瘤分割的新型框架,基于Segment Anything Model(SAM)。
- 采用了两步训练协议:首先微调patch嵌入层处理整个多参数MRI数据,然后融入视觉转换器以捕获切片间的相关性。
- GBT-SAM在成人胶质瘤数据集上取得了最佳性能,Dice分数为93.54%。
- GBT-SAM展现出对脑膜瘤、儿童胶质瘤和撒哈拉以南胶质瘤数据集的稳健泛化能力。
- GBT-SAM使用的可训练参数少于650万,具有高效性。
- GBT-SAM的方法可以在公开代码和模型中找到,网址为:https://github.com/vpulab/med-sam-brain。
- GBT-SAM框架的引入为医学图像分割领域提供了一种新的、高效的解决方案。
点此查看论文截图

WeakMedSAM: Weakly-Supervised Medical Image Segmentation via SAM with Sub-Class Exploration and Prompt Affinity Mining
Authors:Haoran Wang, Lian Huai, Wenbin Li, Lei Qi, Xingqun Jiang, Yinghuan Shi
We have witnessed remarkable progress in foundation models in vision tasks. Currently, several recent works have utilized the segmenting anything model (SAM) to boost the segmentation performance in medical images, where most of them focus on training an adaptor for fine-tuning a large amount of pixel-wise annotated medical images following a fully supervised manner. In this paper, to reduce the labeling cost, we investigate a novel weakly-supervised SAM-based segmentation model, namely WeakMedSAM. Specifically, our proposed WeakMedSAM contains two modules: 1) to mitigate severe co-occurrence in medical images, a sub-class exploration module is introduced to learn accurate feature representations. 2) to improve the quality of the class activation maps, our prompt affinity mining module utilizes the prompt capability of SAM to obtain an affinity map for random-walk refinement. Our method can be applied to any SAM-like backbone, and we conduct experiments with SAMUS and EfficientSAM. The experimental results on three popularly-used benchmark datasets, i.e., BraTS 2019, AbdomenCT-1K, and MSD Cardiac dataset, show the promising results of our proposed WeakMedSAM. Our code is available at https://github.com/wanghr64/WeakMedSAM.
在视觉任务的基础模型中,我们看到了显著的进步。目前,一些最新的工作利用分割任何模型(SAM)来提高医学图像的分割性能,其中大多数工作都集中在以完全监督的方式训练适配器,对大量像素级注释的医学图像进行微调。为了降低标注成本,我们在本文中研究了一种新型的基于弱监督的SAM分割模型,即WeakMedSAM。具体来说,我们提出的WeakMedSAM包含两个模块:1)为了缓解医学图像中严重的共发生问题,引入了子类探索模块来学习准确的特征表示。2)为了提高类激活图的质量,我们的提示亲和力挖掘模块利用SAM的提示能力来获得用于随机游走细化的亲和力图。我们的方法可以应用于任何SAM类似的backbone,我们在SAMUS和EfficientSAM上进行了实验。在三个常用的基准数据集(即BraTS 2019、AbdomenCT-1K和MSD Cardiac数据集)上的实验结果表明,我们提出的WeakMedSAM具有广阔的应用前景。我们的代码可访问于:https://github.com/wanghr64/WeakMedSAM。
论文及项目相关链接
Summary
该论文提出了一种基于弱监督的SAM(分段任何模型)医疗图像分割模型——WeakMedSAM,以降低标注成本。它通过子类别探索模块学习准确的特征表示,并改进了类别激活图的准确性。在SAMUS和EfficientSAM等模型的实验证明其有效性。该模型在BraTS 2019、AbdomenCT-1K和MSD心脏数据集上的实验结果证明了其潜力。代码已公开在GitHub上。
Key Takeaways
- 该论文提出一种基于弱监督的SAM医疗图像分割模型WeakMedSAM,旨在降低标注成本。
- WeakMedSAM包含两个模块:子类探索模块用于学习准确的特征表示,并改进类别激活图的准确性。
- 该模型采用随机游走细化技术,通过SAM的提示能力获取亲和力图。
- WeakMedSAM可应用于任何SAM类似的主干网络,如SAMUS和EfficientSam。
- 实验结果证明了WeakMedSAM在BraTS 2019、AbdomenCT-1K和MSD心脏数据集上的有效性。
点此查看论文截图

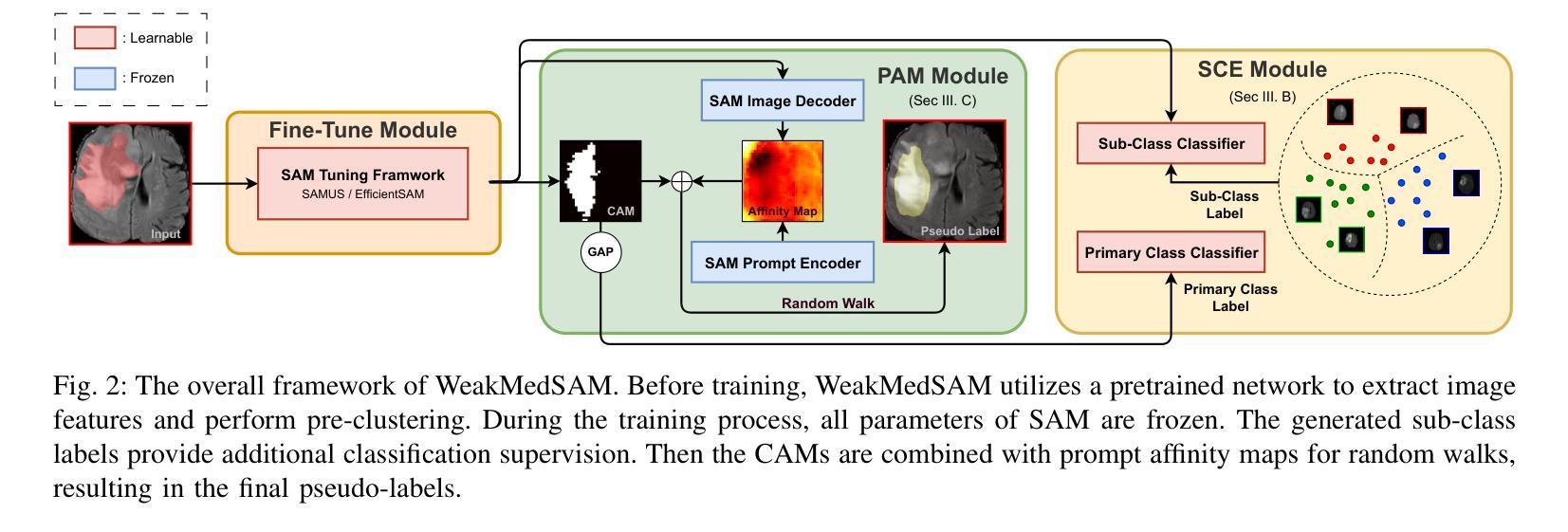
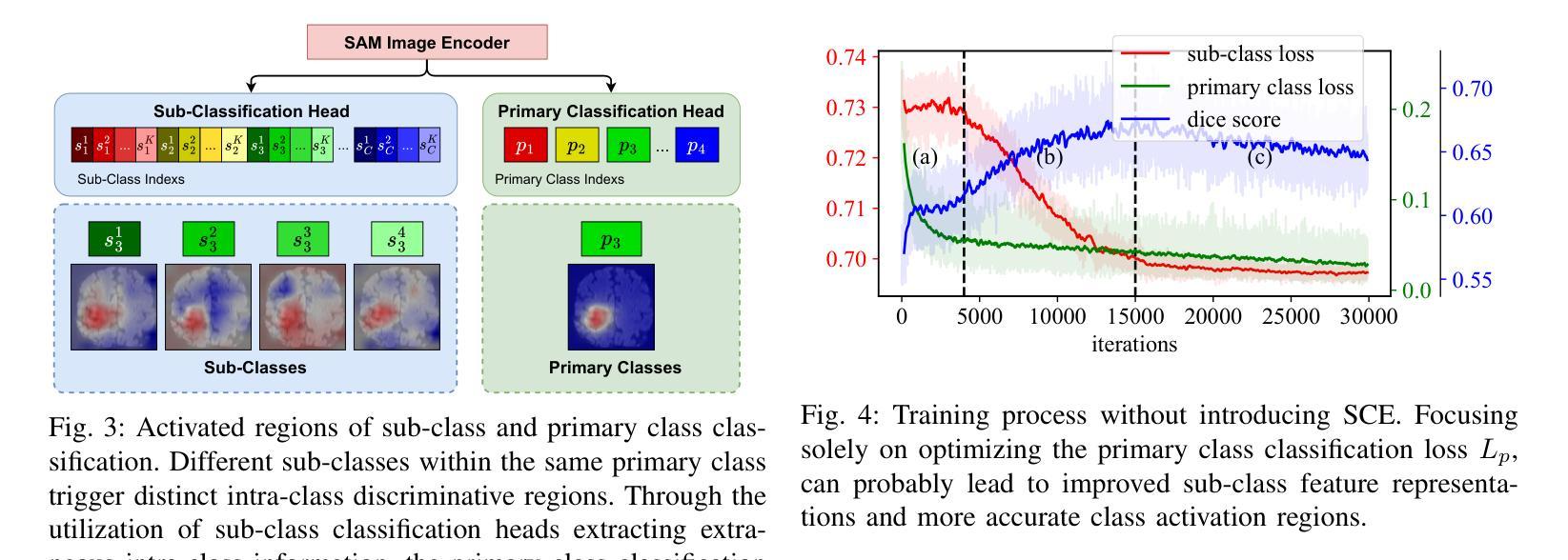
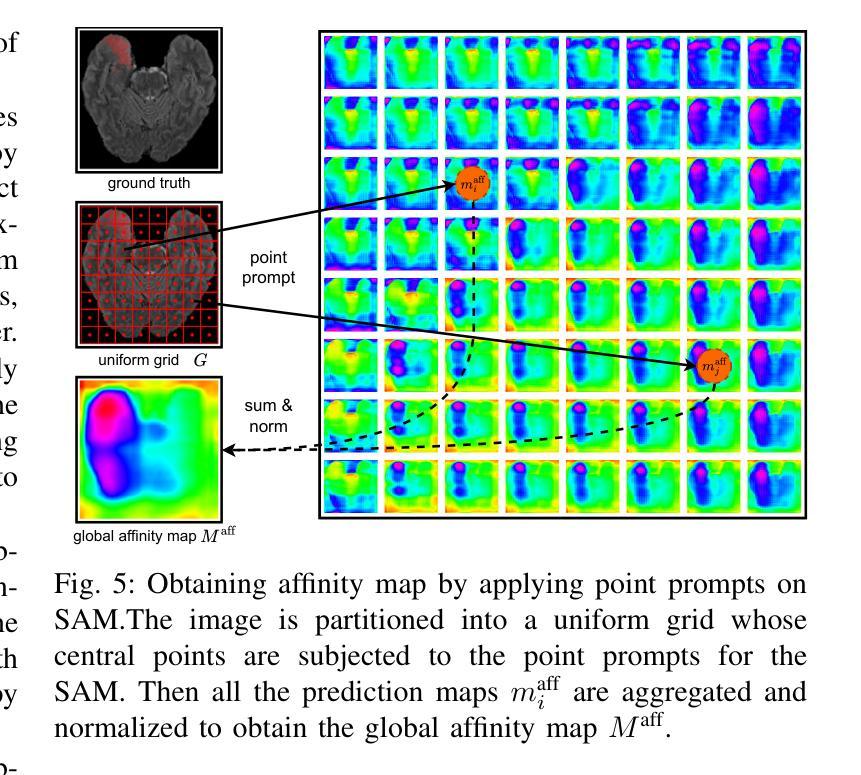
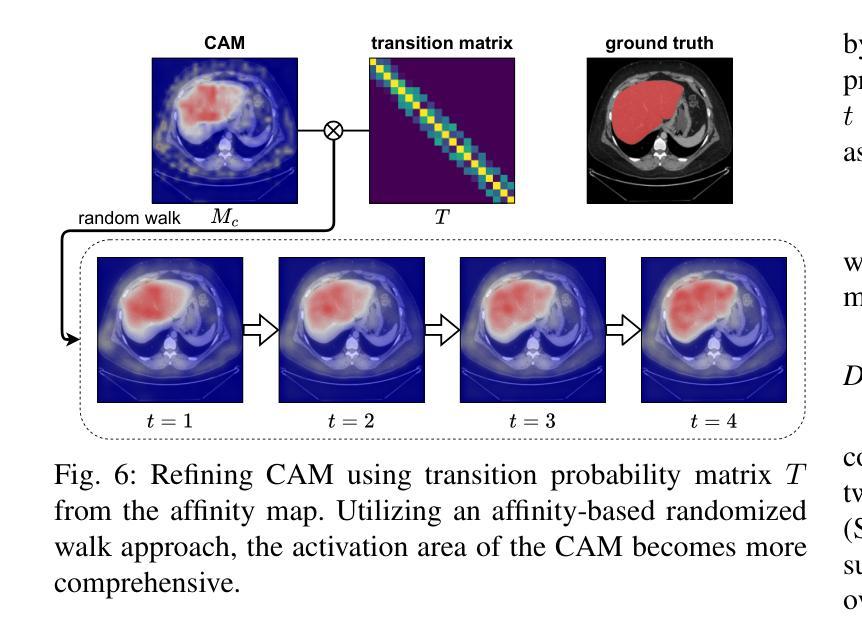
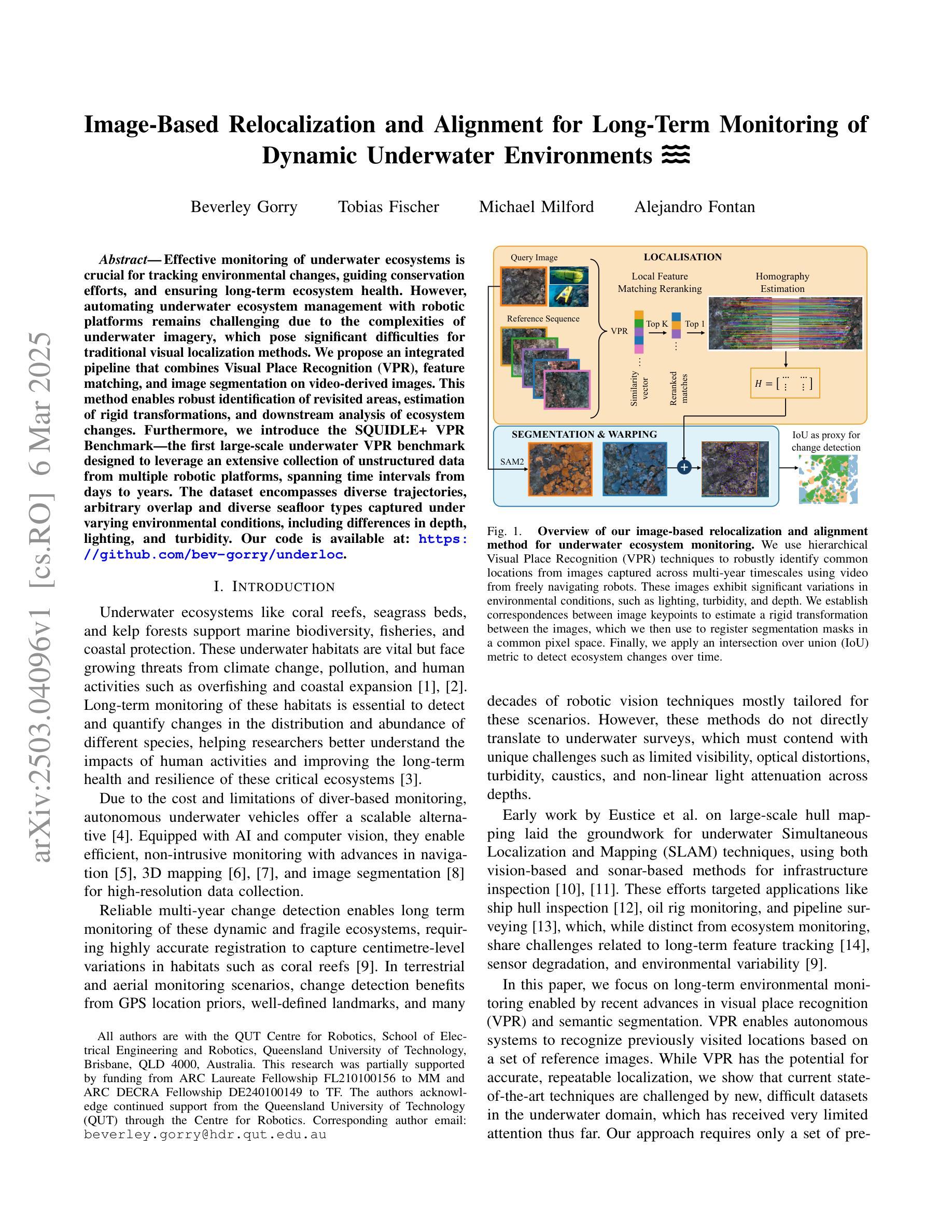
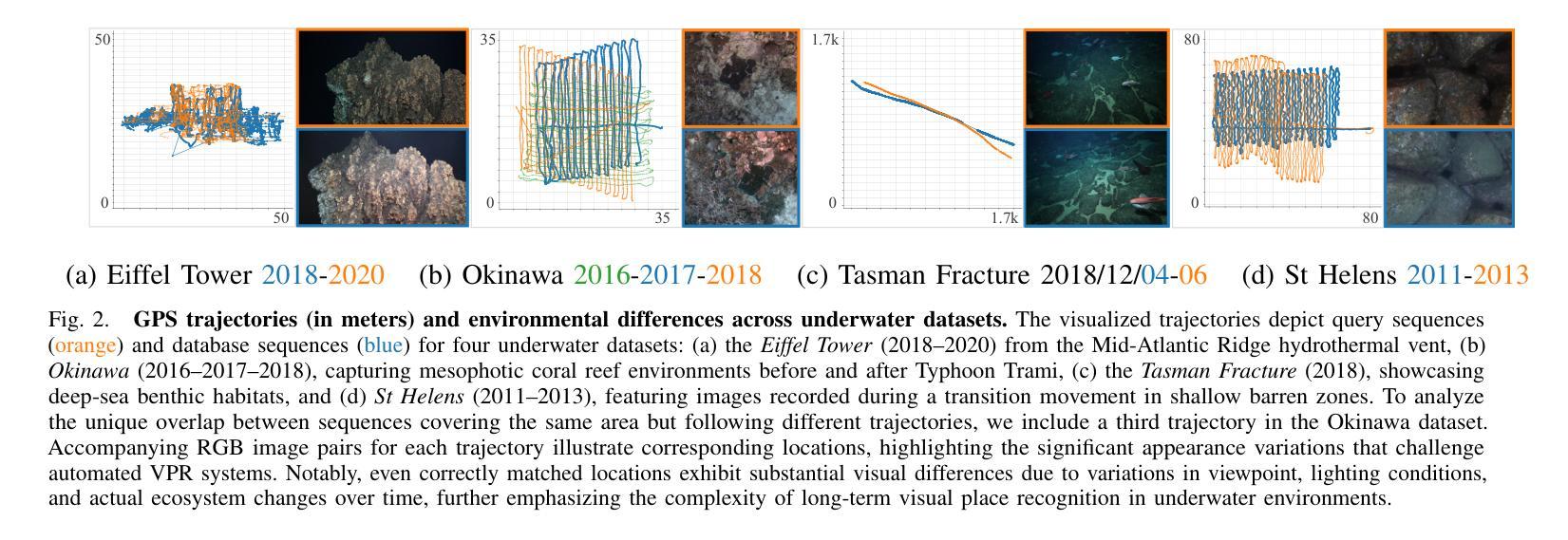
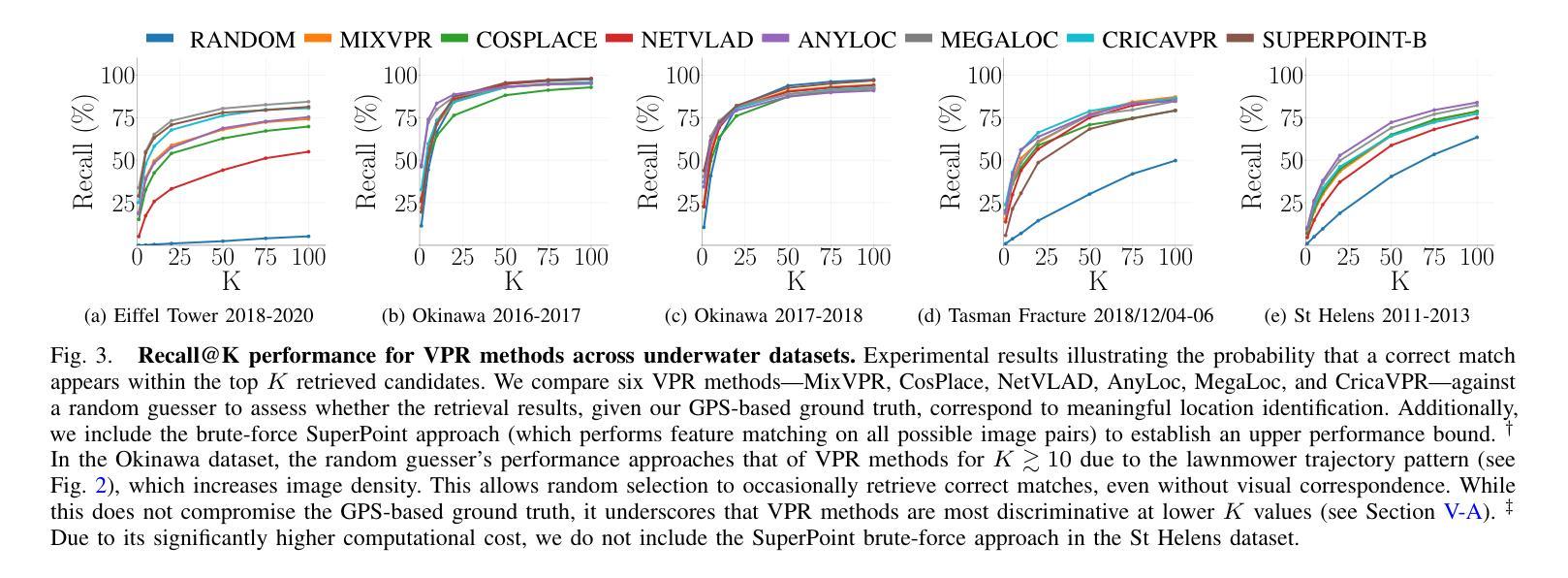
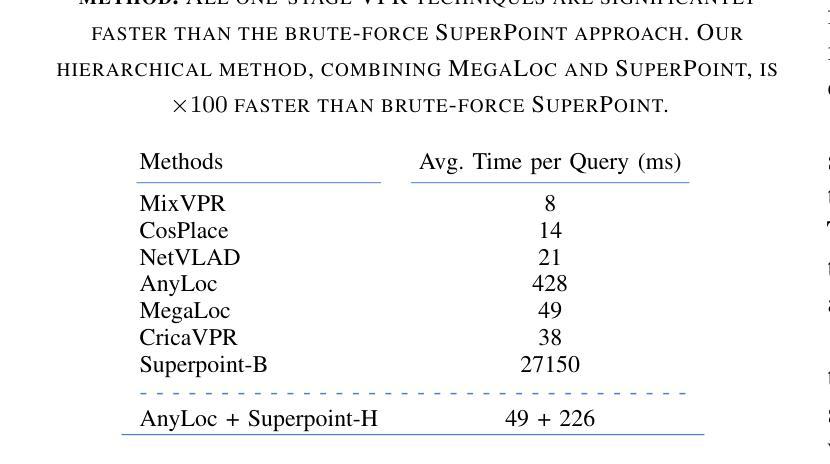
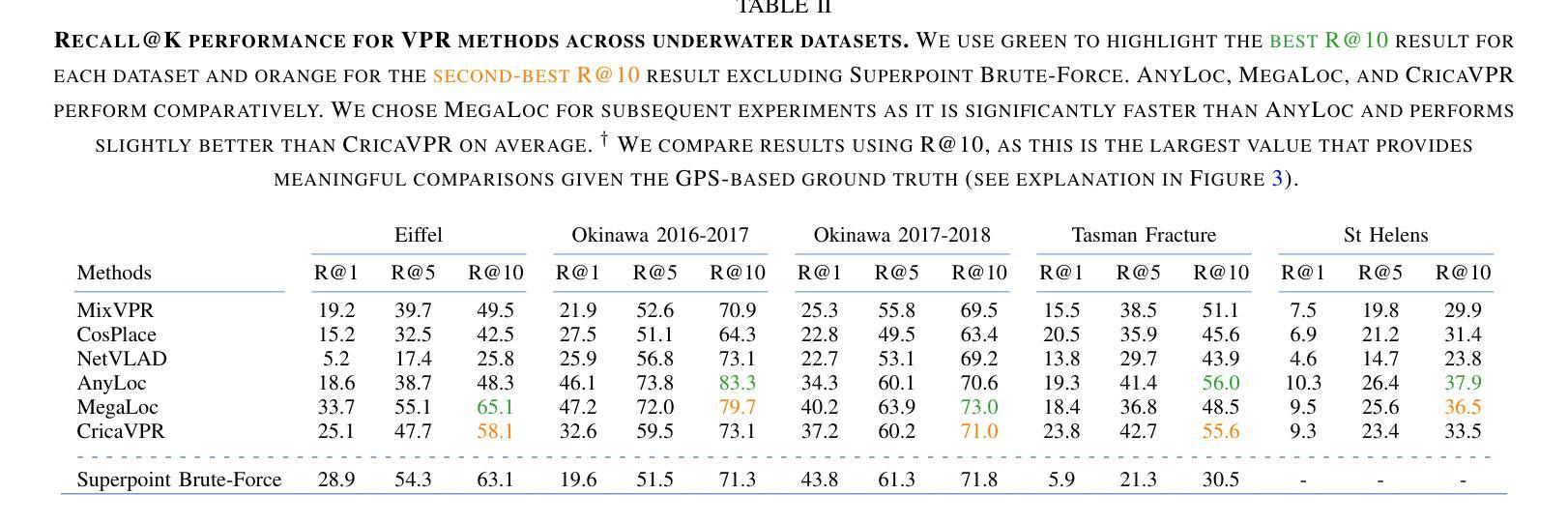
Image-Based Relocalization and Alignment for Long-Term Monitoring of Dynamic Underwater Environments
Authors:Beverley Gorry, Tobias Fischer, Michael Milford, Alejandro Fontan
Effective monitoring of underwater ecosystems is crucial for tracking environmental changes, guiding conservation efforts, and ensuring long-term ecosystem health. However, automating underwater ecosystem management with robotic platforms remains challenging due to the complexities of underwater imagery, which pose significant difficulties for traditional visual localization methods. We propose an integrated pipeline that combines Visual Place Recognition (VPR), feature matching, and image segmentation on video-derived images. This method enables robust identification of revisited areas, estimation of rigid transformations, and downstream analysis of ecosystem changes. Furthermore, we introduce the SQUIDLE+ VPR Benchmark-the first large-scale underwater VPR benchmark designed to leverage an extensive collection of unstructured data from multiple robotic platforms, spanning time intervals from days to years. The dataset encompasses diverse trajectories, arbitrary overlap and diverse seafloor types captured under varying environmental conditions, including differences in depth, lighting, and turbidity. Our code is available at: https://github.com/bev-gorry/underloc
对水下生态系统进行有效监测对于跟踪环境变化、指导保护工作和确保生态系统长期健康至关重要。然而,由于水下图像的复杂性,使用机器人平台自动管理水下生态系统仍然是一个挑战,这给传统的视觉定位方法带来了巨大的困难。我们提出了一种结合视觉定位识别(VPR)、特征匹配和基于视频图像分割方法的综合流程。该方法能够实现稳健的回访区域识别、刚性转换估计以及生态系统变化的下游分析。此外,我们还推出了SQUIDLE+ VPR Benchmark——这是第一个大规模水下VPR基准测试,旨在利用来自多个机器人平台的海量非结构化数据集合,时间间隔从几天到几年不等。数据集包含多种轨迹、任意重叠和不同环境条件下的各种海底类型(包括深度、光照和浊度的差异)。我们的代码可从以下网址获取:https://github.com/bev-gorry/underloc。
论文及项目相关链接
Summary
本文提出一种结合视觉定位识别(VPR)、特征匹配和视频图像分割技术的方法,实现对水下生态系统的自动化监测和管理。通过该方法的实施,能够准确识别回访区域、估算刚性变换以及分析生态系统变化。此外,本文还介绍了首个大规模水下VPR基准测试集——SQUIDLE+ VPR Benchmark,该数据集包含来自多个机器人平台的丰富非结构化数据,跨越数日乃至数年的时间段。数据集涵盖了多种轨迹、任意重叠和不同海底类型等环境条件下的数据差异,包括深度、光照和浊度等。
Key Takeaways
- 有效监测水下生态系统对于跟踪环境变化、指导保护工作和确保生态系统长期健康至关重要。
- 水下成像的复杂性给传统视觉定位方法带来了挑战。
- 提出了一种结合视觉定位识别(VPR)、特征匹配和视频图像分割的集成方法,用于稳健地识别回访区域、估算刚性变换和分析生态系统变化。
- 介绍了首个大规模水下VPR基准测试集——SQUIDLE+ VPR Benchmark。
- 该数据集包含来自多个机器人平台的丰富非结构化数据,跨越不同环境条件下的数据差异。
- 数据集涵盖多种轨迹和任意重叠情况,为水下生态系统管理的自动化提供了重要资源。
点此查看论文截图
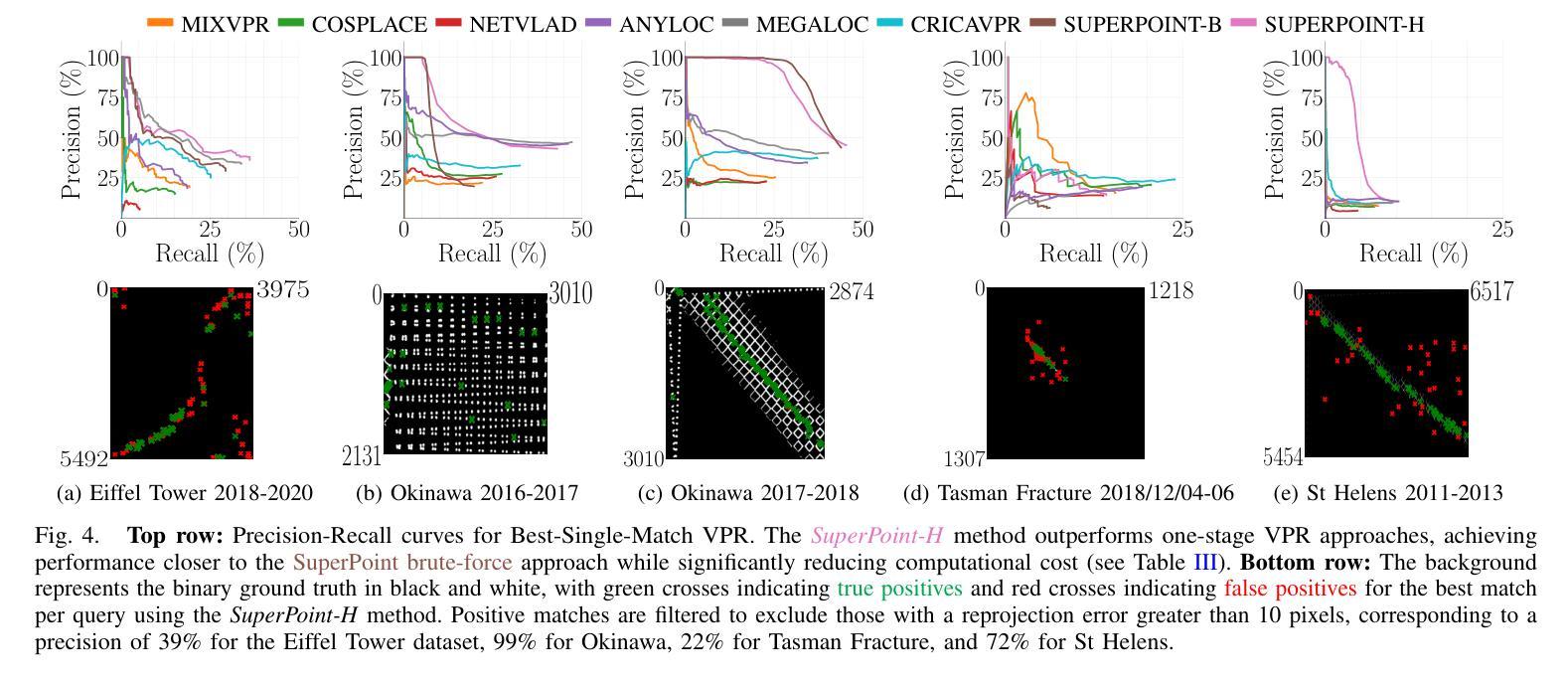
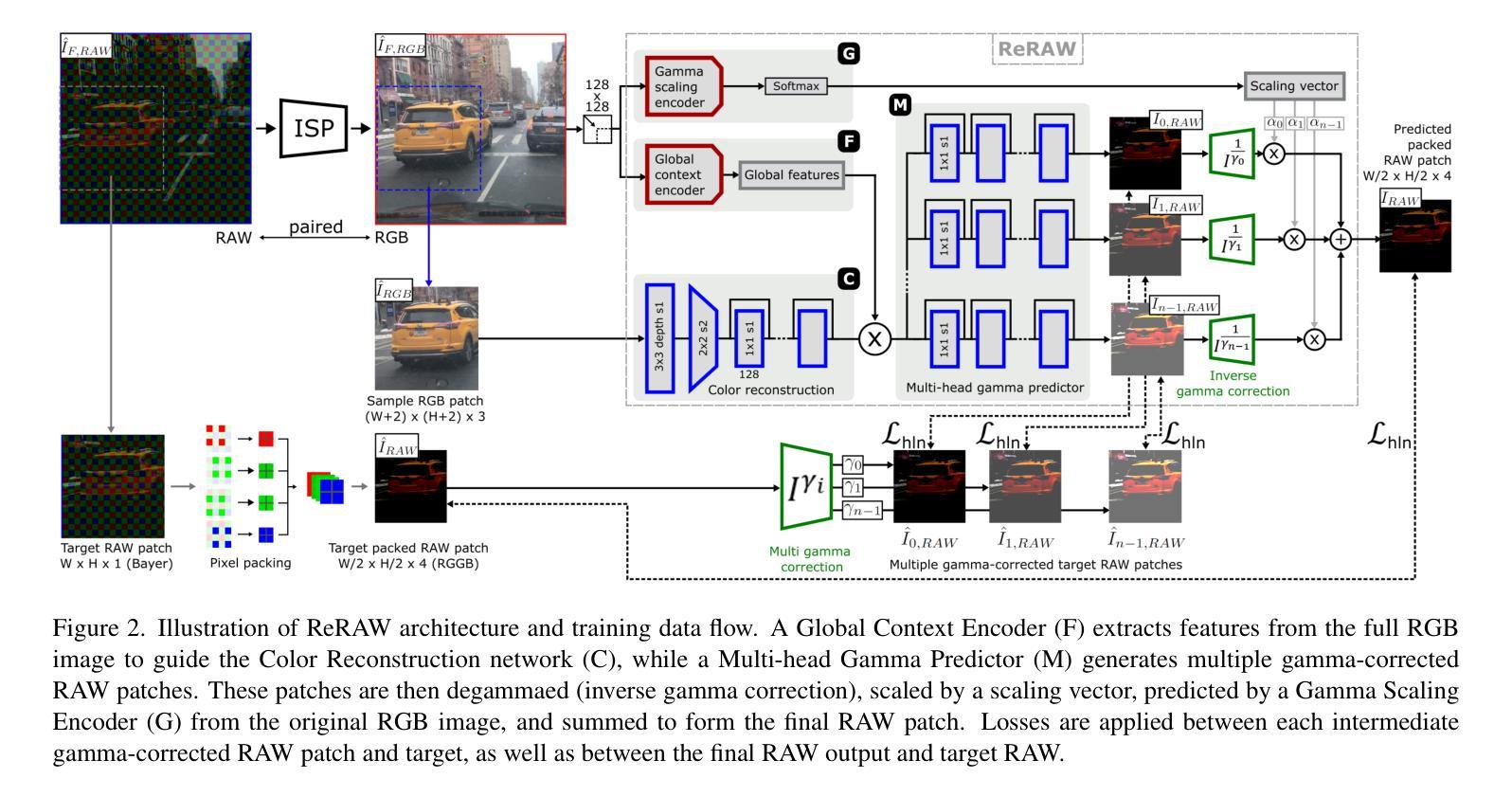
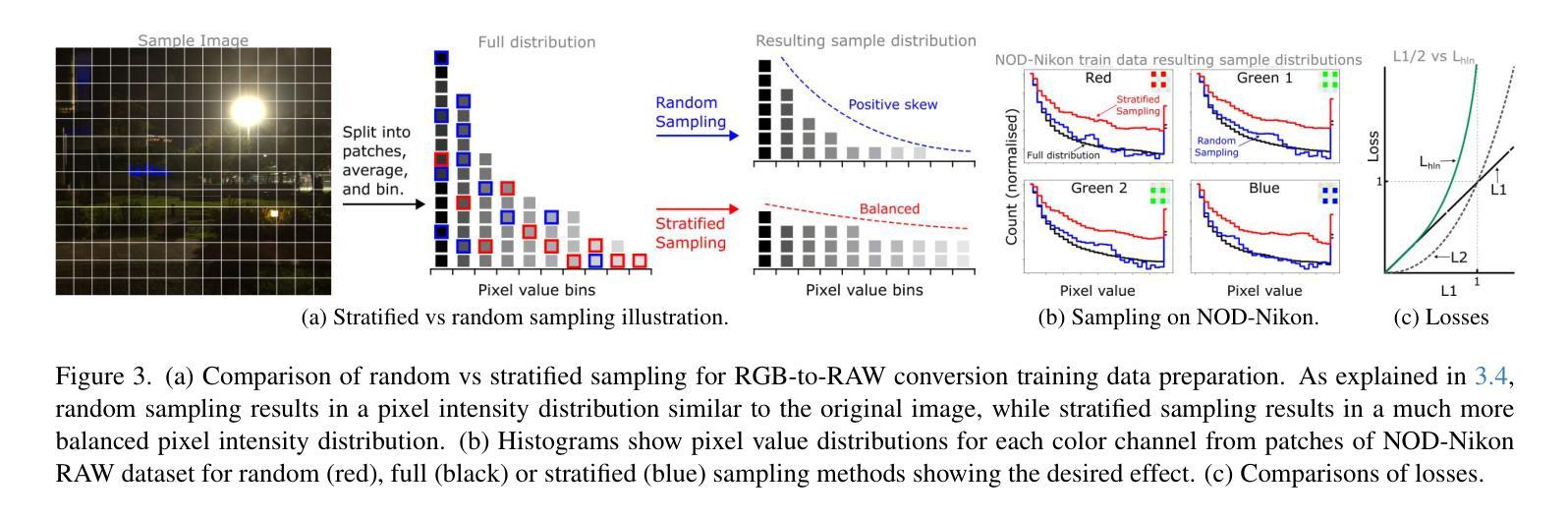
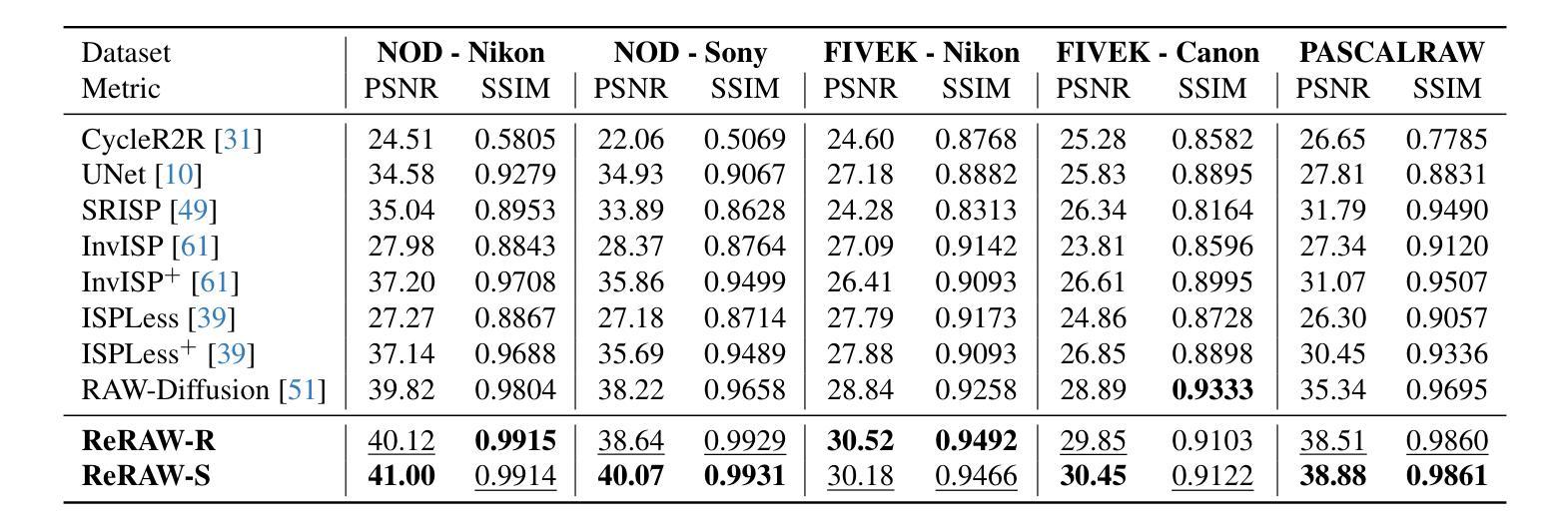
ReRAW: RGB-to-RAW Image Reconstruction via Stratified Sampling for Efficient Object Detection on the Edge
Authors:Radu Berdan, Beril Besbinar, Christoph Reinders, Junji Otsuka, Daisuke Iso
Edge-based computer vision models running on compact, resource-limited devices benefit greatly from using unprocessed, detail-rich RAW sensor data instead of processed RGB images. Training these models, however, necessitates large labeled RAW datasets, which are costly and often impractical to obtain. Thus, converting existing labeled RGB datasets into sensor-specific RAW images becomes crucial for effective model training. In this paper, we introduce ReRAW, an RGB-to-RAW conversion model that achieves state-of-the-art reconstruction performance across five diverse RAW datasets. This is accomplished through ReRAW’s novel multi-head architecture predicting RAW image candidates in gamma space. The performance is further boosted by a stratified sampling-based training data selection heuristic, which helps the model better reconstruct brighter RAW pixels. We finally demonstrate that pretraining compact models on a combination of high-quality synthetic RAW datasets (such as generated by ReRAW) and ground-truth RAW images for downstream tasks like object detection, outperforms both standard RGB pipelines, and RAW fine-tuning of RGB-pretrained models for the same task.
基于边缘的计算机视觉模型在紧凑、资源受限的设备上运行,从使用未经处理、细节丰富的RAW传感器数据而不是经过处理的RGB图像中获益匪浅。然而,训练这些模型需要大量的标记RAW数据集,这既昂贵又往往不切实际。因此,将现有的标记RGB数据集转换为特定传感器RAW图像对于有效模型训练变得至关重要。在本文中,我们介绍了ReRAW,一种RGB到RAW的转换模型,它在五个不同的RAW数据集上实现了最先进的重建性能。这是通过ReRAW的新型多头架构在伽马空间中预测RAW图像候选物来实现的。通过分层采样为基础的训练数据选择启发式方法,性能得到了进一步提升,这有助于模型更好地重建较亮的RAW像素。最后,我们证明在高质量合成RAW数据集(如由ReRAW生成)和真实RAW图像的组合上预训练紧凑模型,用于下游任务(如对象检测),优于标准的RGB管道和相同任务的RAW微调RGB预训练模型。
论文及项目相关链接
PDF Accepted at CVPR 2025
Summary
采用无处理、细节丰富的RAW传感器数据,在资源有限的设备上运行的边缘计算视觉模型相较于使用已处理的RGB图像受益匪浅。然而,训练这些模型需要大量标记的RAW数据集,这成本高昂并不切实际。因此,将现有标记的RGB数据集转换为特定传感器RAW图像变得至关重要。本文介绍了一种RGB转RAW转换模型ReRAW,其在五个不同的RAW数据集上实现了最先进的重建性能。其创新的多头架构能够在伽马空间预测RAW图像候选者,从而实现这一目标。此外,基于分层采样的训练数据选择启发式策略进一步提高了性能,帮助模型更好地重建较亮的RAW像素。最后证明,在下游任务如目标检测上,使用高质量合成RAW数据集(如由ReRAW生成)和真实RAW图像预训练紧凑模型,相较于标准RGB管道和RGB预训练模型的RAW微调,表现更优异。
Key Takeaways
- 使用未处理的RAW传感器数据对边缘计算视觉模型进行训练在资源受限的设备上具有显著优势。
- 训练这类模型需要大量标记的RAW数据集,但其获取成本高昂且实际操作困难。
- ReRAW模型通过RGB转RAW转换实现了先进的重建性能,借助其多头架构在伽马空间预测RAW图像。
- 分层采样启发式的训练数据选择策略增强了模型的性能,特别是在重建亮像素方面。
- 高质量合成RAW数据集与真实RAW图像的结合使用对于预训练紧凑模型至关重要。
- 预训练模型在目标检测等下游任务上的表现超越了标准RGB管道和RGB预训练模型的RAW微调。
点此查看论文截图


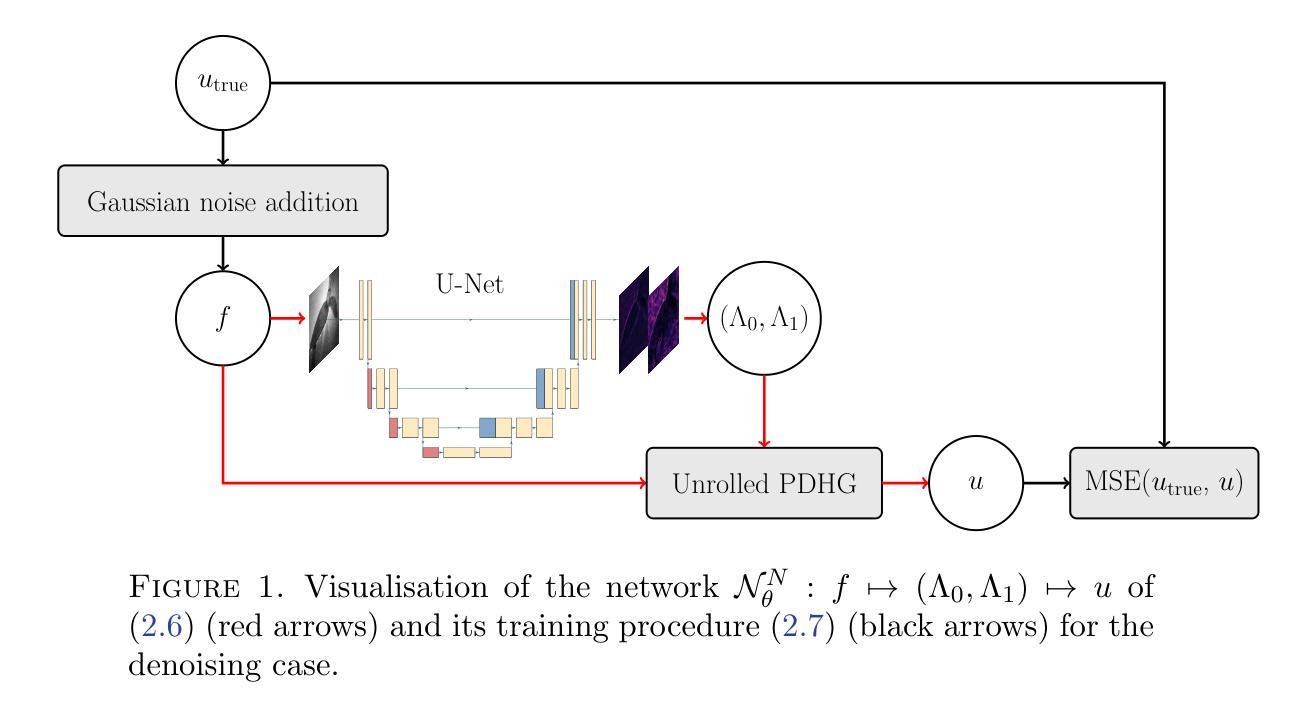
Deep unrolling for learning optimal spatially varying regularisation parameters for Total Generalised Variation
Authors:Thanh Trung Vu, Andreas Kofler, Kostas Papafitsoros
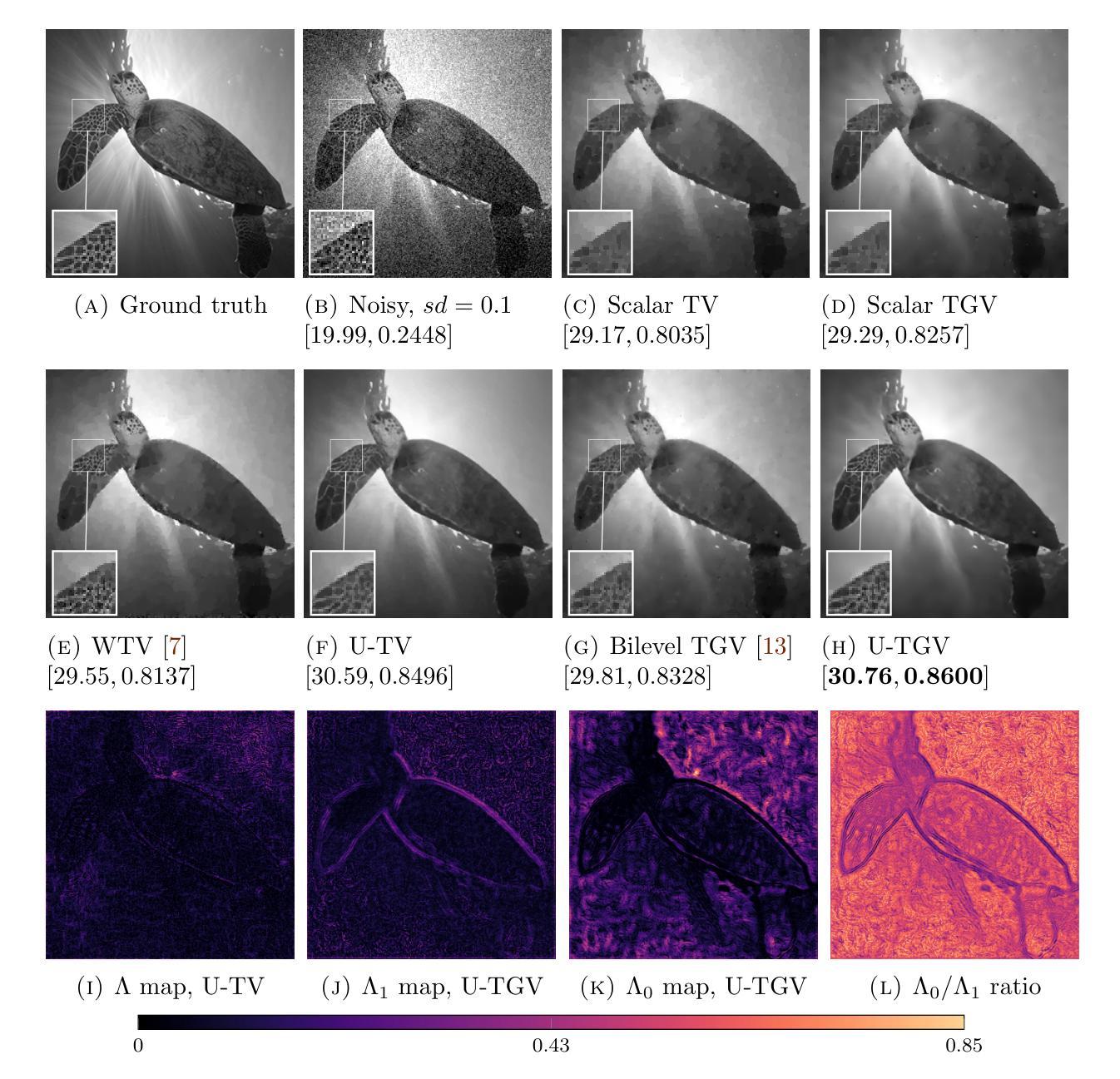
We extend a recently introduced deep unrolling framework for learning spatially varying regularisation parameters in inverse imaging problems to the case of Total Generalised Variation (TGV). The framework combines a deep convolutional neural network (CNN) inferring the two spatially varying TGV parameters with an unrolled algorithmic scheme that solves the corresponding variational problem. The two subnetworks are jointly trained end-to-end in a supervised fashion and as such the CNN learns to compute those parameters that drive the reconstructed images as close to the ground truth as possible. Numerical results in image denoising and MRI reconstruction show a significant qualitative and quantitative improvement compared to the best TGV scalar parameter case as well as to other approaches employing spatially varying parameters computed by unsupervised methods. We also observe that the inferred spatially varying parameter maps have a consistent structure near the image edges, asking for further theoretical investigations. In particular, the parameter that weighs the first-order TGV term has a triple-edge structure with alternating high-low-high values whereas the one that weighs the second-order term attains small values in a large neighbourhood around the edges.
我们将最近引入的深度展开框架扩展到总广义变化(TGV)的情况,用于学习反成像问题中的空间变化正则化参数。该框架结合了深度卷积神经网络(CNN),用于推断两个空间变化的TGV参数,以及一个解决相应变分问题的展开算法方案。两个子网络以监督方式端对端联合训练,因此CNN学习计算那些参数,以尽可能接近真实情况的方式重建图像。在图像去噪和MRI重建方面的数值结果表明,与最佳TGV标量参数情况以及其他采用空间变化参数的方法相比,无论在定性和定量上均有显著改善。这些空间变化的参数图在图像边缘附近具有一致的结构,需要进一步的理论研究。特别是,权重一阶TGV项的参数字映射具有高低高的三重边缘结构,而权重二阶项的参数在边缘附近的大范围内达到较小值。
论文及项目相关链接
Summary
本文将深度展开框架扩展到总广义变化(TGV)的情况,结合深度卷积神经网络(CNN)推断两个空间变化的TGV参数与一个解决相应变分问题的展开算法方案。两个子网络以监督方式进行端到端的联合训练,使得CNN学习计算使重建图像尽可能接近真实值的参数。图像去噪和MRI重建的数值结果显示,与最佳TGV标量参数情况以及其他采用空间变化参数的计算方法相比,本文方法有显著的定性和定量改进。
Key Takeaways
- 深度展开框架被扩展到总广义变化(TGV)的情况。
- 结合CNN推断空间变化的TGV参数与展开算法方案。
- 两个子网络以监督方式进行端到端的联合训练。
- CNN学习计算使重建图像尽可能接近真实值的参数。
- 数值结果显示该方法在图像去噪和MRI重建上有显著改进。
- 推断出的空间变化参数地图在图像边缘附近有一致的结构。
- 一阶TGV参数的权重具有三边结构,而二阶TGV参数的权重在边缘周围达到小值。
点此查看论文截图

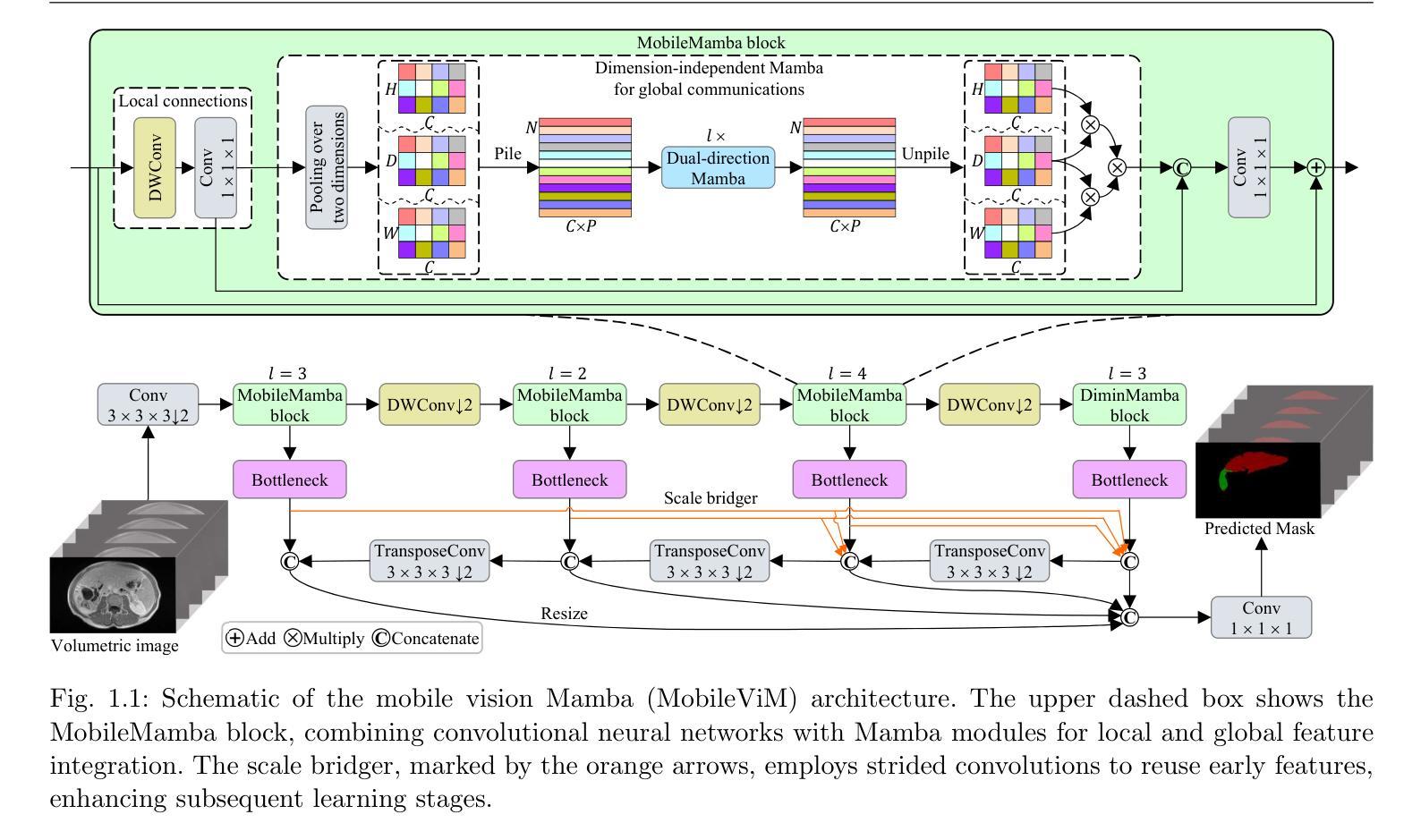
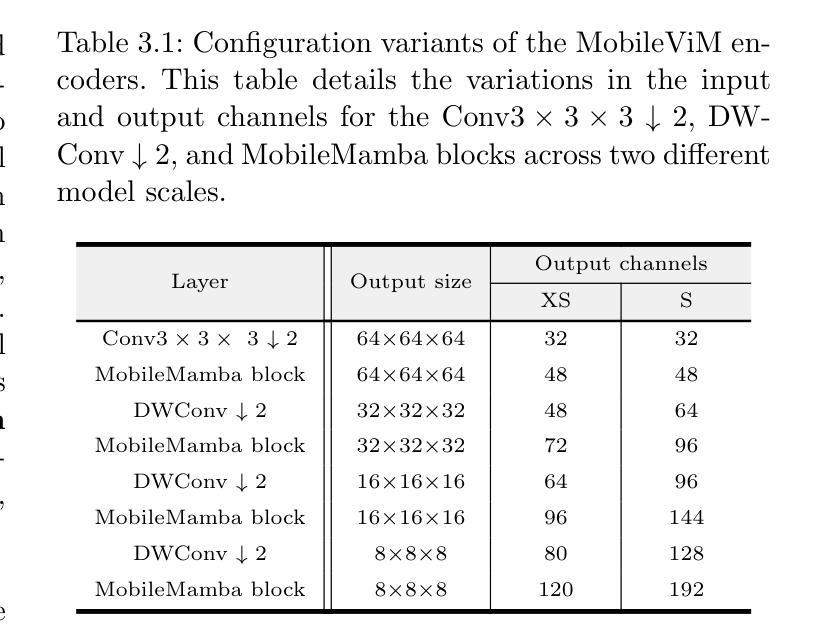
MobileViM: A Light-weight and Dimension-independent Vision Mamba for 3D Medical Image Analysis
Authors:Wei Dai, Jun Liu
Efficient evaluation of three-dimensional (3D) medical images is crucial for diagnostic and therapeutic practices in healthcare. Recent years have seen a substantial uptake in applying deep learning and computer vision to analyse and interpret medical images. Traditional approaches, such as convolutional neural networks (CNNs) and vision transformers (ViTs), face significant computational challenges, prompting the need for architectural advancements. Recent efforts have led to the introduction of novel architectures like the ``Mamba’’ model as alternative solutions to traditional CNNs or ViTs. The Mamba model excels in the linear processing of one-dimensional data with low computational demands. However, Mamba’s potential for 3D medical image analysis remains underexplored and could face significant computational challenges as the dimension increases. This manuscript presents MobileViM, a streamlined architecture for efficient segmentation of 3D medical images. In the MobileViM network, we invent a new dimension-independent mechanism and a dual-direction traversing approach to incorporate with a vision-Mamba-based framework. MobileViM also features a cross-scale bridging technique to improve efficiency and accuracy across various medical imaging modalities. With these enhancements, MobileViM achieves segmentation speeds exceeding 90 frames per second (FPS) on a single graphics processing unit (i.e., NVIDIA RTX 4090). This performance is over 24 FPS faster than the state-of-the-art deep learning models for processing 3D images with the same computational resources. In addition, experimental evaluations demonstrate that MobileViM delivers superior performance, with Dice similarity scores reaching 92.72%, 86.69%, 80.46%, and 77.43% for PENGWIN, BraTS2024, ATLAS, and Toothfairy2 datasets, respectively, which significantly surpasses existing models.
在医疗保健领域,对三维(3D)医学图像的有效评估对于诊断和治疗实践至关重要。近年来,深度学习和计算机视觉在医学图像分析和解释方面的应用取得了显著进展。传统方法,如卷积神经网络(CNNs)和视觉转换器(ViTs),面临着重大的计算挑战,这促使了架构发展的必要性。最近的努力导致了诸如“Mamba”模型等新架构的引入,作为传统CNN或ViTs的替代解决方案。Mamba模型在处理一维数据时具有出色的线性处理能力且计算需求较低。然而,Mamba在三维医学图像分析方面的潜力尚未得到充分探索,随着维度的增加,可能会面临重大的计算挑战。本文提出了MobileViM,这是一个用于高效分割三维医学图像的简化架构。在MobileViM网络中,我们发明了一种新的与维度无关的机制以及一种双向遍历方法与基于视觉Mamba的框架相结合。MobileViM还采用跨尺度桥梁技术,以提高各种医学成像模式的有效性和准确性。通过这些增强功能,MobileViM在单个图形处理单元(即NVIDIA RTX 4090)上实现了超过每秒90帧(FPS)的分割速度。这一性能比使用相同计算资源的处理三维图像的最先进深度学习模型的帧率快24 FPS以上。此外,实验评估表明,MobileViM具有卓越的性能,在PENGWIN、BraTS2024、ATLAS和Toothfairy2数据集上的Dice相似度得分分别达到92.72%、86.69%、80.46%和77.43%,显著超过了现有模型。
论文及项目相关链接
PDF The corresponding author disagrees with the manuscript submitted to arXiv
Summary
本文介绍了一种针对三维医学图像分析的新型高效架构MobileViM。它通过引入维度独立机制、双向遍历方法和跨尺度桥接技术,实现了快速而精确的医学图像分割。相较于其他顶尖深度学习模型,MobileViM在单一GPU上的运行速度提升了超过24FPS,并且在多个数据集上的Dice相似度得分表现出卓越性能。
Key Takeaways
- MobileViM是一种针对三维医学图像分析的高效架构。
- MobileViM引入了维度独立机制,适应不同维度的数据处理。
- 通过双向遍历方法,MobileViM提升了图像处理的效率。
- 跨尺度桥接技术提高了MobileViM在不同医学影像模态下的性能和准确性。
- MobileViM实现了超过90FPS的分割速度,在单一GPU上的性能超越现有模型。
- MobileViM在多个数据集上的Dice相似度得分表现出卓越性能。
点此查看论文截图

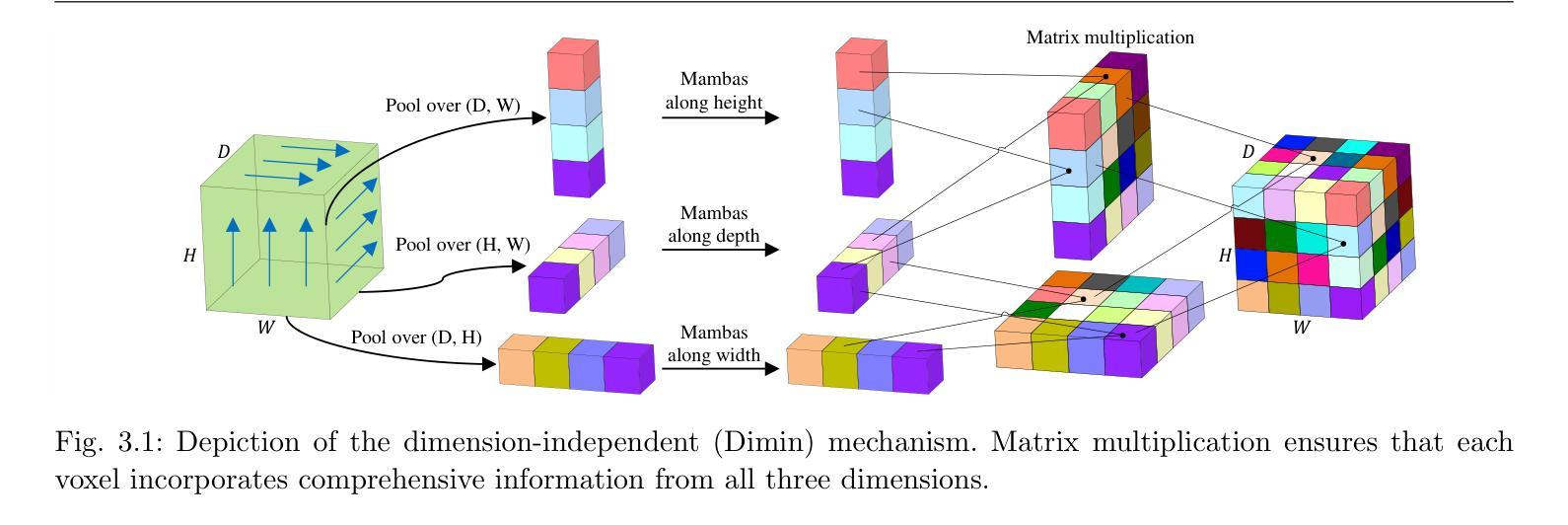
Game-Theoretic Defenses for Robust Conformal Prediction Against Adversarial Attacks in Medical Imaging
Authors:Rui Luo, Jie Bao, Zhixin Zhou, Chuangyin Dang
Adversarial attacks pose significant threats to the reliability and safety of deep learning models, especially in critical domains such as medical imaging. This paper introduces a novel framework that integrates conformal prediction with game-theoretic defensive strategies to enhance model robustness against both known and unknown adversarial perturbations. We address three primary research questions: constructing valid and efficient conformal prediction sets under known attacks (RQ1), ensuring coverage under unknown attacks through conservative thresholding (RQ2), and determining optimal defensive strategies within a zero-sum game framework (RQ3). Our methodology involves training specialized defensive models against specific attack types and employing maximum and minimum classifiers to aggregate defenses effectively. Extensive experiments conducted on the MedMNIST datasets, including PathMNIST, OrganAMNIST, and TissueMNIST, demonstrate that our approach maintains high coverage guarantees while minimizing prediction set sizes. The game-theoretic analysis reveals that the optimal defensive strategy often converges to a singular robust model, outperforming uniform and simple strategies across all evaluated datasets. This work advances the state-of-the-art in uncertainty quantification and adversarial robustness, providing a reliable mechanism for deploying deep learning models in adversarial environments.
对抗性攻击对深度学习模型的可靠性和安全性构成重大威胁,特别是在医疗成像等关键领域。本文介绍了一个新型框架,该框架将合规性预测与博弈理论防御策略相结合,以提高模型对已知和未知对抗性扰动的鲁棒性。我们解决了三个主要的研究问题:在已知攻击下构建有效且高效的合规性预测集(RQ1),通过保守阈值化确保在未知攻击下的覆盖率(RQ2),以及在零和博弈框架内确定最佳防御策略(RQ3)。我们的方法包括针对特定攻击类型训练专门的防御模型,并使用最大和最小分类器有效地聚合防御措施。在MedMNIST数据集(包括PathMNIST、OrganAMNIST和TissueMNIST)上进行的广泛实验表明,我们的方法在保持高覆盖率保证的同时,最小化了预测集的大小。博弈理论分析表明,最佳防御策略通常会收敛到一个单一的稳健模型,在所有评估的数据集上,其性能都优于均匀和简单策略。这项工作在不确定性的量化和对抗稳健性方面达到了最新水平,为在敌对环境中部署深度学习模型提供了可靠的机制。
论文及项目相关链接
Summary
本论文提出一种新型框架,结合保形预测和游戏理论防御策略,提高深度学习模型对抗已知和未知对抗性扰动的稳健性。研究解决三个主要问题:在已知攻击下构建有效且高效的保形预测集、通过保守阈值化确保未知攻击下的覆盖,以及在零和博弈框架内确定最佳防御策略。实验证明,该方法在保持高覆盖保证的同时减小了预测集大小。游戏理论分析显示,最佳防御策略通常收敛于单一稳健模型,在所有评估数据集上表现优于均匀和简单策略。
Key Takeaways
- 对抗攻击对深度学习模型的可靠性和安全性构成重大威胁,特别是在医疗成像等关键领域。
- 论文提出结合保形预测和游戏理论防御策略的新型框架,增强模型对抗已知和未知对抗性扰动的稳健性。
- 研究解决三个主要问题:构建保形预测集、确保未知攻击下的覆盖以及确定最佳防御策略。
- 通过在MedMNIST数据集(包括PathMNIST、OrganAMNIST和TissueMNIST)上进行广泛实验,证明该方法在保持高覆盖保证的同时减小预测集大小。
- 游戏理论分析显示,最佳防御策略通常收敛于单一稳健模型,该模型在多个数据集上的性能优于均匀和简单策略。
- 该方法提高了不确定性量化的水平,并增强了对抗性环境下的模型稳健性。
点此查看论文截图

LDPM: Towards undersampled MRI reconstruction with MR-VAE and Latent Diffusion Prior
Authors:Xingjian Tang, Jingwei Guan, Linge Li, Ran Shi, Youmei Zhang, Mengye Lyu, Li Yan
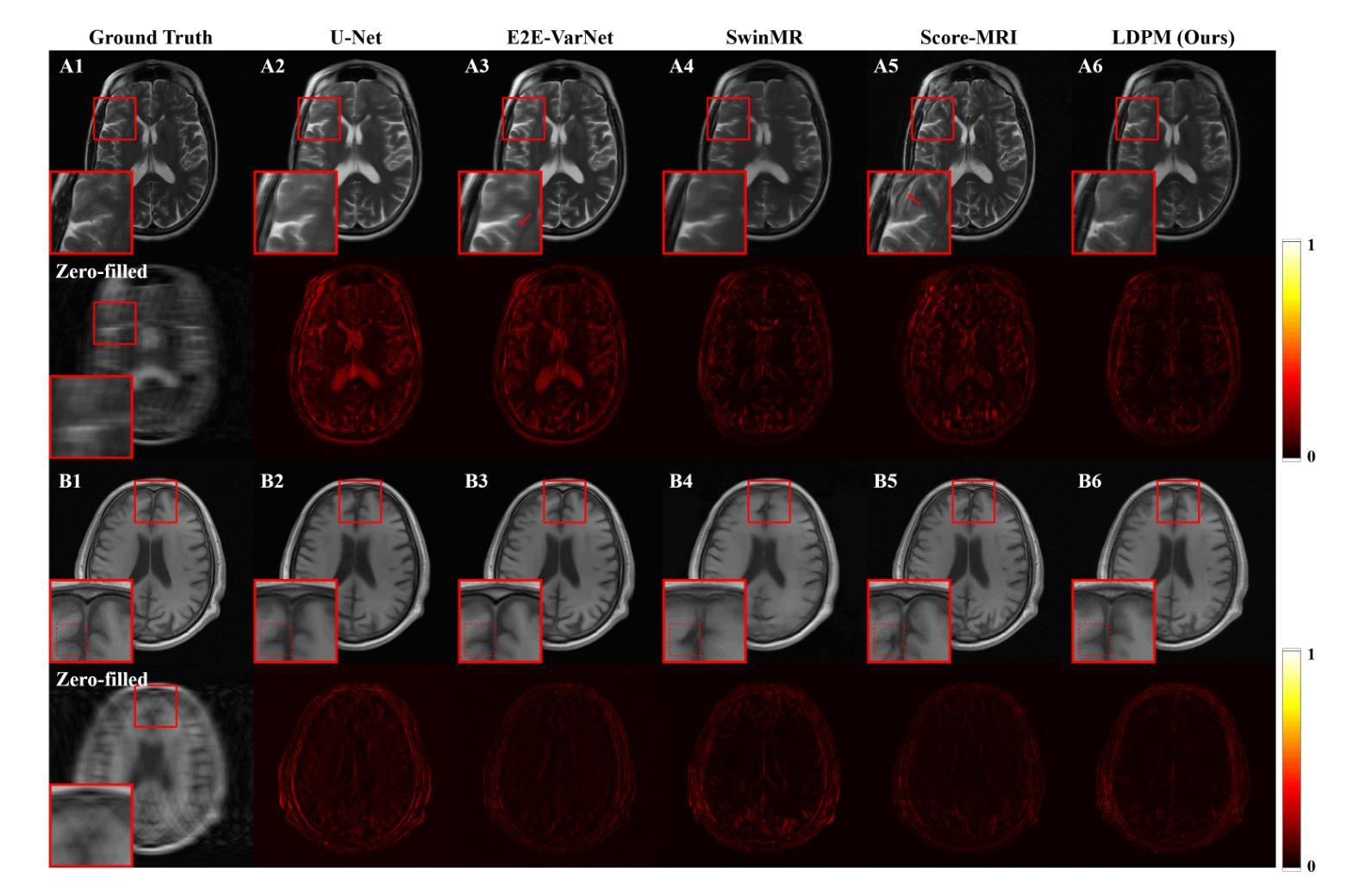
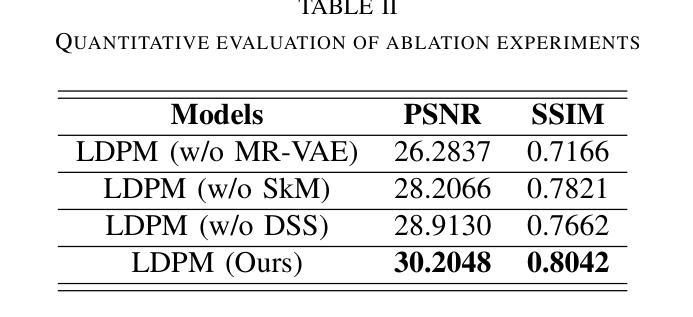
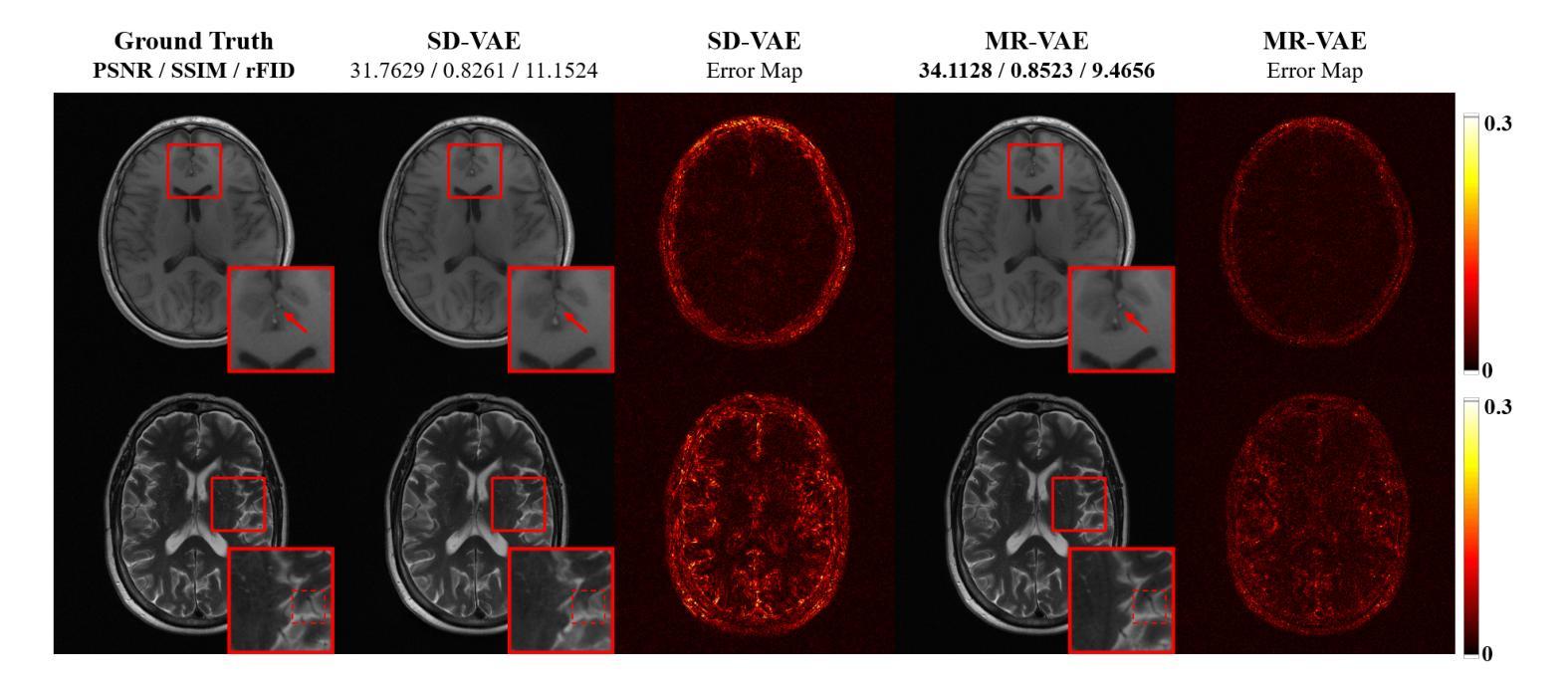
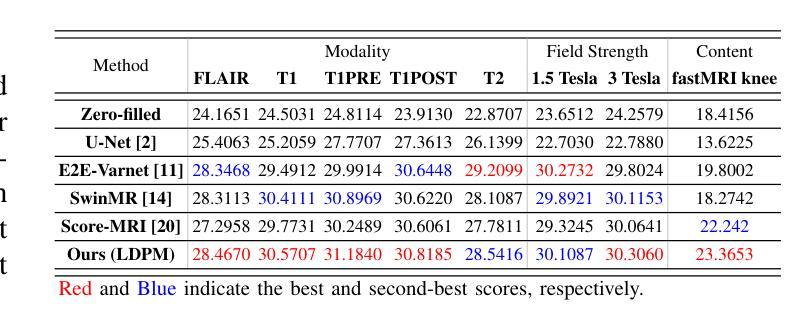
Diffusion models, as powerful generative models, have found a wide range of applications and shown great potential in solving image reconstruction problems. Some works attempted to solve MRI reconstruction with diffusion models, but these methods operate directly in pixel space, leading to higher computational costs for optimization and inference. Latent diffusion models, pre-trained on natural images with rich visual priors, are expected to solve the high computational cost problem in MRI reconstruction by operating in a lower-dimensional latent space. However, direct application to MRI reconstruction faces three key challenges: (1) absence of explicit control mechanisms for medical fidelity, (2) domain gap between natural images and MR physics, and (3) undefined data consistency in latent space. To address these challenges, a novel Latent Diffusion Prior-based undersampled MRI reconstruction (LDPM) method is proposed. Our LDPM framework addresses these challenges by: (1) a sketch-guided pipeline with a two-step reconstruction strategy, which balances perceptual quality and anatomical fidelity, (2) an MRI-optimized VAE (MR-VAE), which achieves an improvement of approximately 3.92 dB in PSNR for undersampled MRI reconstruction compared to that with SD-VAE \cite{sd}, and (3) Dual-Stage Sampler, a modified version of spaced DDPM sampler, which enforces high-fidelity reconstruction in the latent space. Experiments on the fastMRI dataset\cite{fastmri} demonstrate the state-of-the-art performance of the proposed method and its robustness across various scenarios. The effectiveness of each module is also verified through ablation experiments.
扩散模型作为强大的生成模型,在图像重建问题中得到了广泛的应用,并显示出巨大的潜力。一些研究尝试使用扩散模型解决MRI重建问题,但这些方法在像素空间直接操作,导致优化和推理的计算成本较高。潜在扩散模型在自然图像上进行预训练,具有丰富的视觉先验,有望通过低维潜在空间解决MRI重建中的高计算成本问题。然而,直接应用于MRI重建面临三个关键挑战:(1)医学保真度的控制机制缺失,(2)自然图像与MR物理之间的领域差距,(3)潜在空间中的数据一致性未定义。为了解决这些挑战,提出了一种基于潜在扩散先验的欠采样MRI重建(LDPM)新方法。我们的LDPM框架通过以下方式应对这些挑战:(1)一个草图引导的流程,采用两步重建策略,平衡感知质量和解剖保真度;(2)一个针对MRI优化的VAE(MR-VAE),在欠采样MRI重建的PSNR上相比SD-VAE \cite{sd}提高了约3.92 dB;(3)双阶段采样器,这是一个改进的间隔DDPM采样器,它在潜在空间中强制高保真重建。在fastMRI数据集\cite{fastmri}上的实验证明了该方法的先进性能和在各种场景中的稳健性。每个模块的有效性也通过消融实验得到了验证。
论文及项目相关链接
Summary
潜在扩散模型在解决MRI重建问题方面具有巨大潜力,但仍面临挑战。新型LDPM方法通过草图引导管道、MRI优化的VAE和双阶段采样器等创新手段,解决了这些问题,并在fastMRI数据集上展现出卓越性能。
Key Takeaways
- 扩散模型在图像重建问题中展现了巨大的潜力。
- 直接在像素空间操作MRI重建导致高计算成本。
- 潜在扩散模型期望通过在低维潜在空间操作来解决高计算成本问题。
- LDPM方法通过草图引导管道、MRI优化的VAE和双阶段采样器解决潜在扩散模型在MRI重建中的挑战。
- LDPM方法在fastMRI数据集上实现了先进性能,并展示了在各种场景下的稳健性。
- LDPM方法通过平衡感知质量和解剖保真度来提高MRI重建的质量。
点此查看论文截图

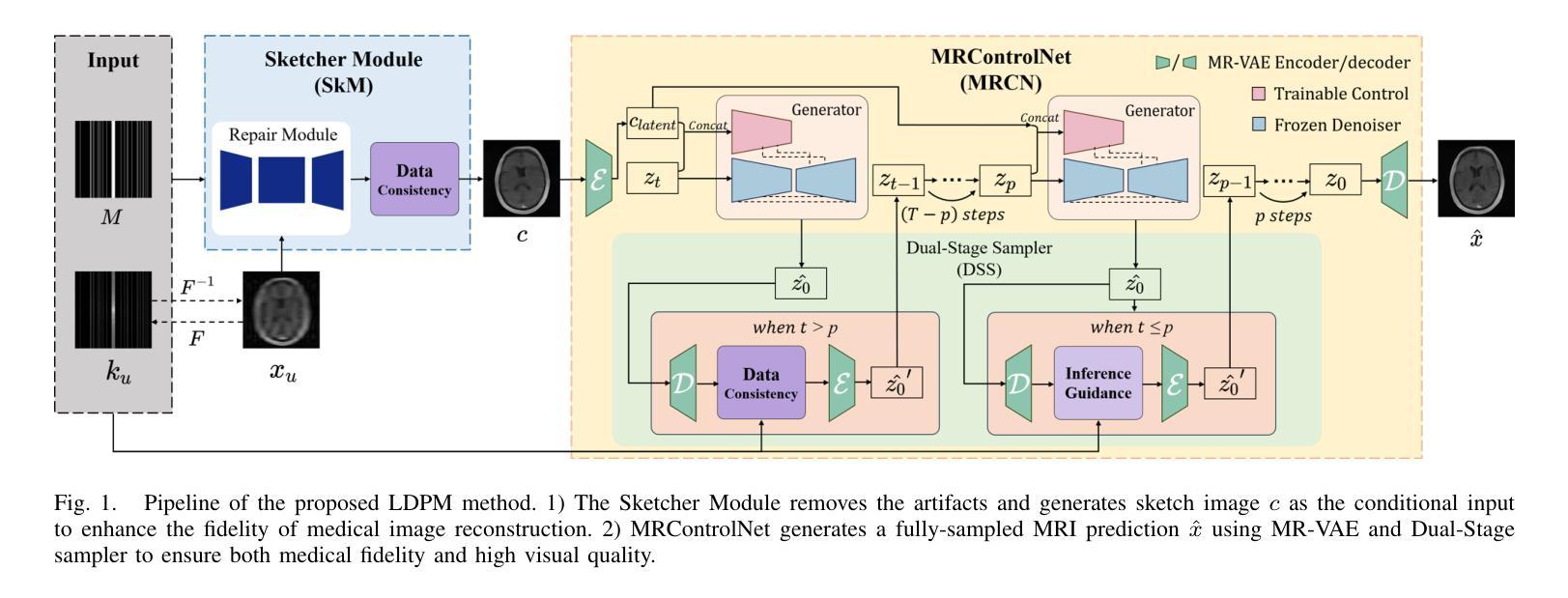
Enhancing Multimodal Medical Image Classification using Cross-Graph Modal Contrastive Learning
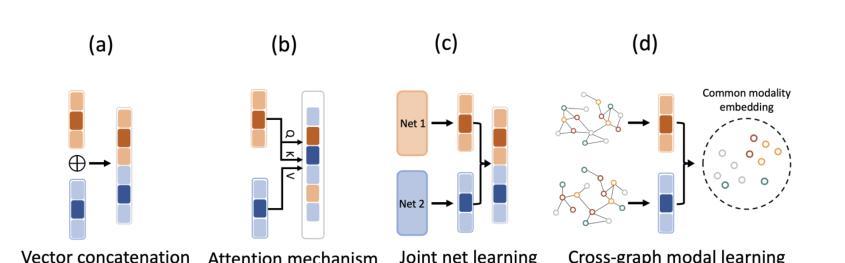
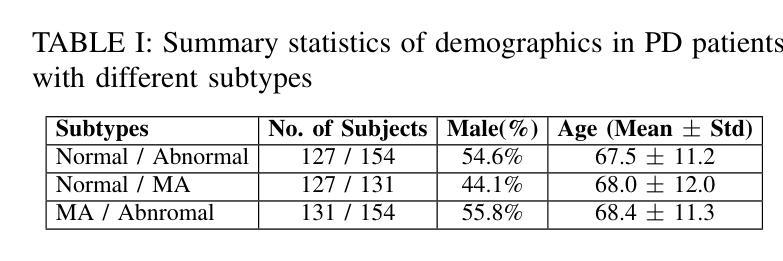
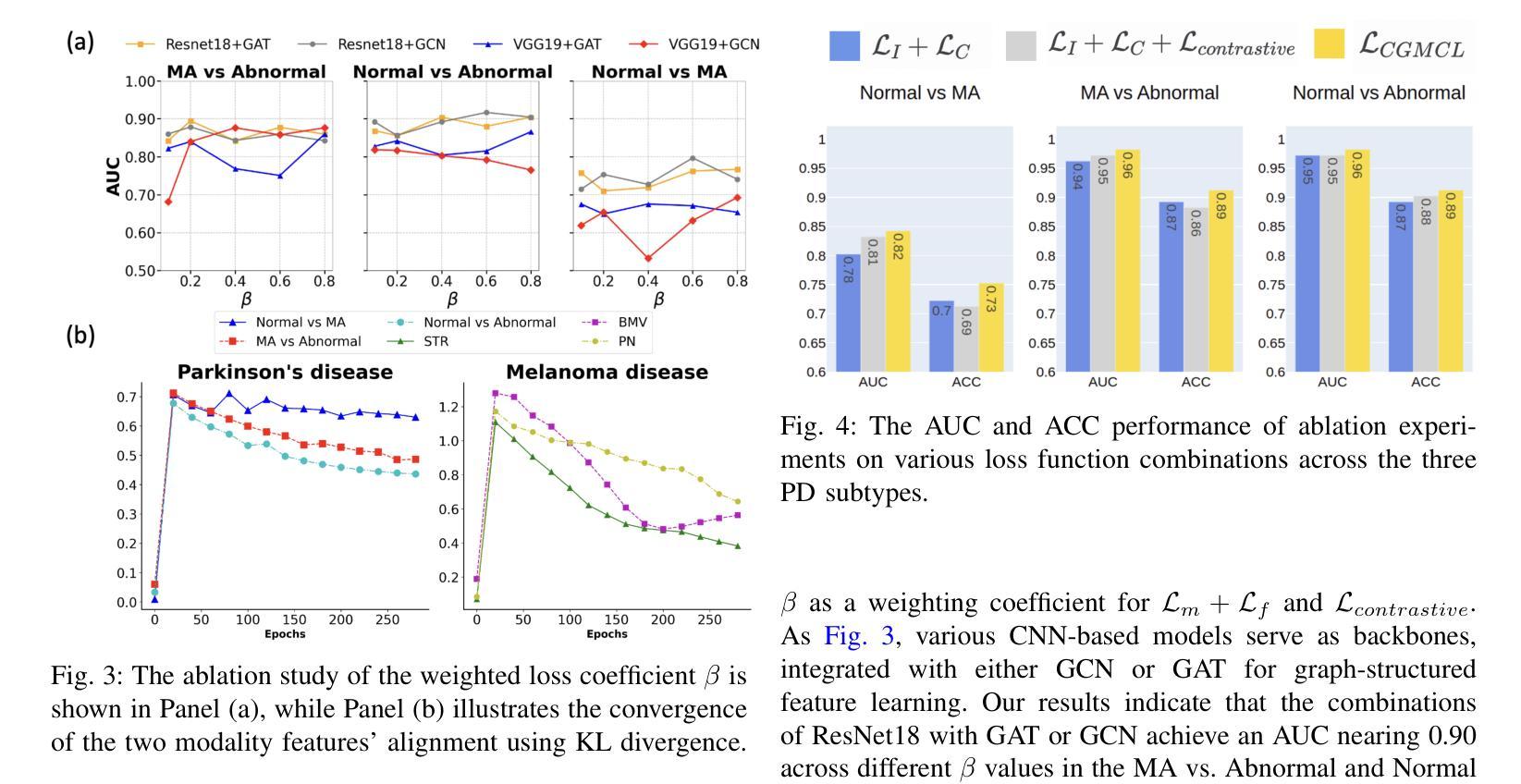
Authors:Jun-En Ding, Chien-Chin Hsu, Chi-Hsiang Chu, Shuqiang Wang, Feng Liu
The classification of medical images is a pivotal aspect of disease diagnosis, often enhanced by deep learning techniques. However, traditional approaches typically focus on unimodal medical image data, neglecting the integration of diverse non-image patient data. This paper proposes a novel Cross-Graph Modal Contrastive Learning (CGMCL) framework for multimodal structured data from different data domains to improve medical image classification. The model effectively integrates both image and non-image data by constructing cross-modality graphs and leveraging contrastive learning to align multimodal features in a shared latent space. An inter-modality feature scaling module further optimizes the representation learning process by reducing the gap between heterogeneous modalities. The proposed approach is evaluated on two datasets: a Parkinson’s disease (PD) dataset and a public melanoma dataset. Results demonstrate that CGMCL outperforms conventional unimodal methods in accuracy, interpretability, and early disease prediction. Additionally, the method shows superior performance in multi-class melanoma classification. The CGMCL framework provides valuable insights into medical image classification while offering improved disease interpretability and predictive capabilities.
医学图像分类是疾病诊断的关键环节,通常可以通过深度学习技术得到增强。然而,传统方法主要关注单模态医学图像数据,忽略了不同非图像患者数据的整合。本文提出了一种新型的跨图模态对比学习(CGMCL)框架,用于从不同数据域的多模态结构化数据,以提高医学图像分类的效果。该模型通过构建跨模态图并利用对比学习,有效地整合了图像和非图像数据,在共享潜在空间中对齐多模态特征。跨模态特征缩放模块进一步优化了表示学习过程,缩小了不同模态之间的差距。所提出的方法在两个数据集上进行了评估:帕金森病(PD)数据集和公共黑色素瘤数据集。结果表明,在准确性、可解释性和早期疾病预测方面,CGMCL优于传统的单模态方法。此外,该方法在多类黑色素瘤分类方面也表现出卓越的性能。CGMCL框架为医学图像分类提供了有价值的见解,同时提高了疾病可解释性和预测能力。
论文及项目相关链接
Summary
本文提出了一种新型的跨图模态对比学习(CGMCL)框架,用于多模态结构化数据的跨域融合,以提高医学图像分类的准确性。该框架通过构建跨模态图并利用对比学习,在共享潜在空间中对齐多模态特征,实现图像与非图像数据的融合。该方法优化了多模态特征的表示学习过程,减少了不同模态之间的差距。在两个数据集上的评估结果证明,与常规的单模态方法相比,CGmcl在准确性、可解释性和早期疾病预测方面表现出优越的性能。特别是在多类黑色素瘤分类方面,CGmcl框架提供了宝贵的医学图像分类洞察力,提高了疾病可解释性和预测能力。
Key Takeaways
- 医学图像分类是疾病诊断的重要环节,而深度学习技术能增强其准确性。
- 传统方法主要关注单模态医学图像数据,忽略了不同非图像患者数据的整合。
- 提出的Cross-Graph Modal Contrastive Learning (CGMCL)框架旨在整合多模态结构化数据,提高医学图像分类效果。
- CGMCL框架通过构建跨模态图,利用对比学习对齐多模态特征,实现图像与非图像数据的融合。
- CGMCL框架优化了多模态特征的表示学习过程,缩小了不同模态间的差距。
- 在帕金森氏病和黑色素瘤数据集上的评估显示,CGmcl在准确性、可解释性和早期疾病预测方面优于传统方法。
点此查看论文截图


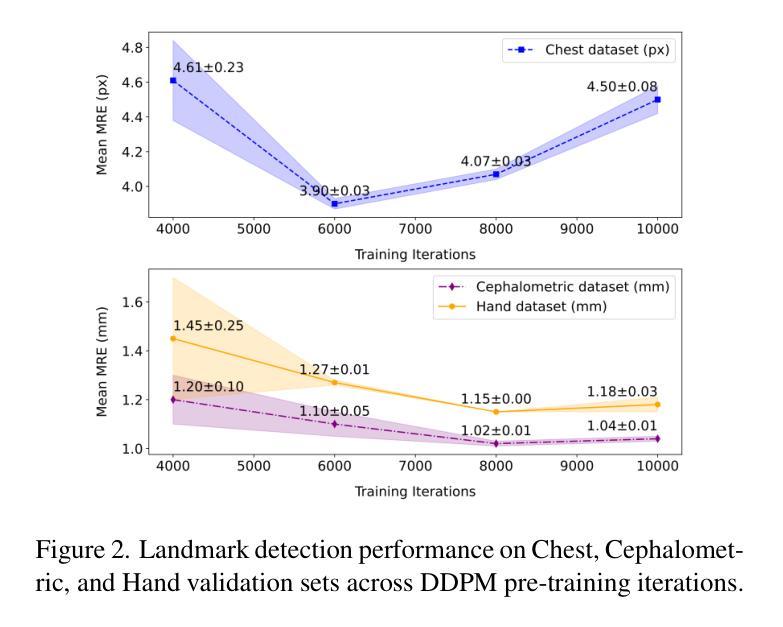
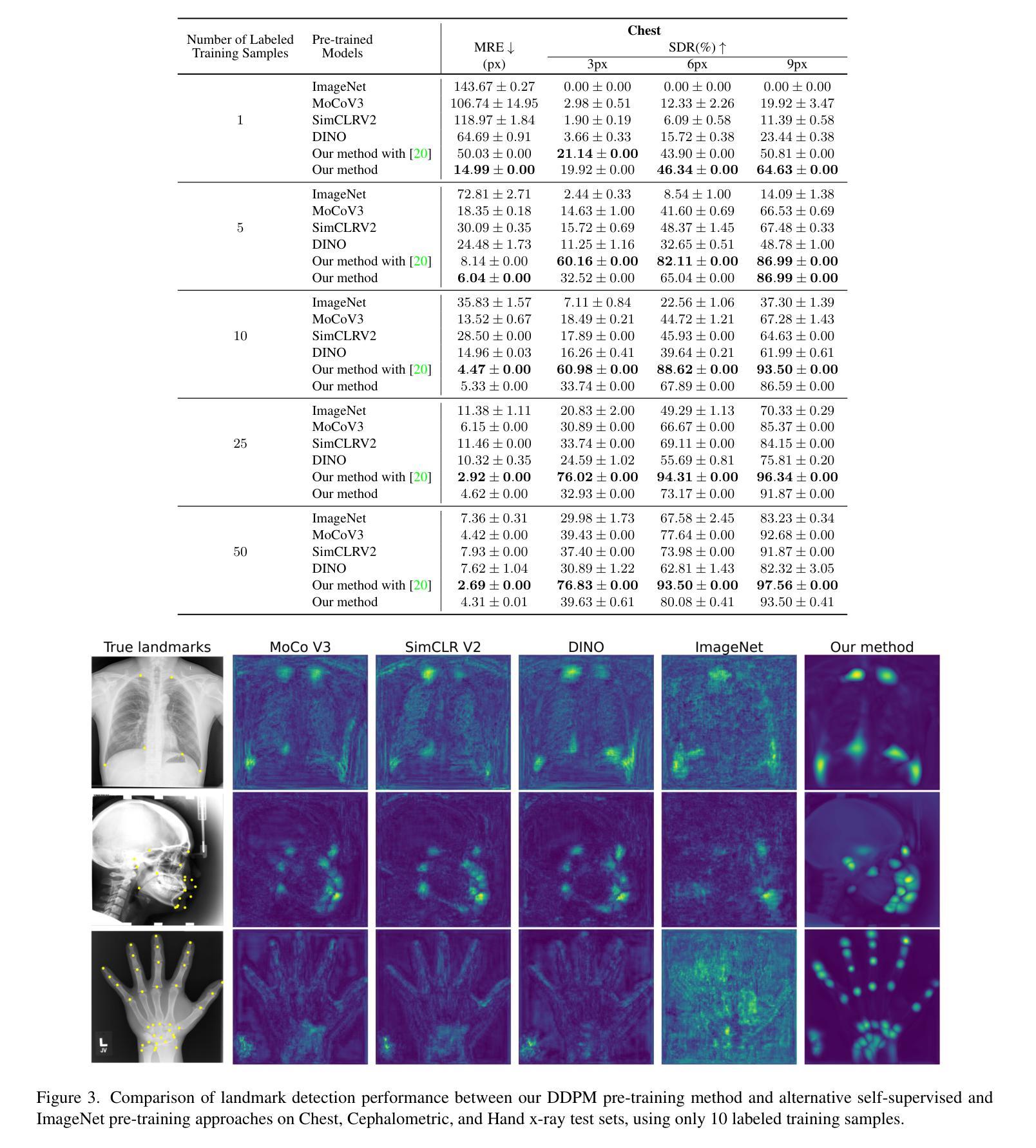
Self-supervised pre-training with diffusion model for few-shot landmark detection in x-ray images
Authors:Roberto Di Via, Francesca Odone, Vito Paolo Pastore
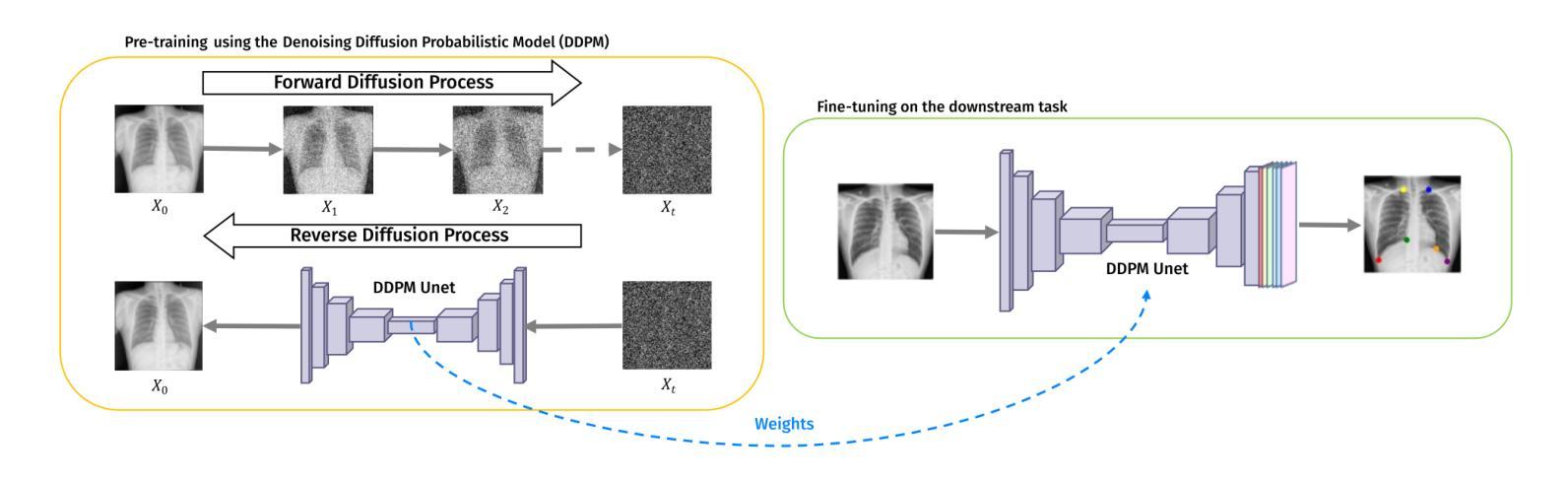
Deep neural networks have been extensively applied in the medical domain for various tasks, including image classification, segmentation, and landmark detection. However, their application is often hindered by data scarcity, both in terms of available annotations and images. This study introduces a novel application of denoising diffusion probabilistic models (DDPMs) to the landmark detection task, specifically addressing the challenge of limited annotated data in x-ray imaging. Our key innovation lies in leveraging DDPMs for self-supervised pre-training in landmark detection, a previously unexplored approach in this domain. This method enables accurate landmark detection with minimal annotated training data (as few as 50 images), surpassing both ImageNet supervised pre-training and traditional self-supervised techniques across three popular x-ray benchmark datasets. To our knowledge, this work represents the first application of diffusion models for self-supervised learning in landmark detection, which may offer a valuable pre-training approach in few-shot regimes, for mitigating data scarcity.
深度神经网络已在医疗领域得到广泛应用,用于各种任务,包括图像分类、分割和地标检测。然而,其应用往往受到数据和注释可用性的限制。本研究引入了去噪扩散概率模型(DDPMs)的新应用,专门解决X射线成像中注释数据有限所带来的挑战。我们的主要创新之处在于,利用DDPMs进行地标检测的自监督预训练,这是该领域之前未被探索的方法。该方法能够在极少的注释训练数据(仅50张图像)的情况下实现准确的地标检测,超越了ImageNet监督预训练和传统自监督技术在三个流行的X射线基准数据集上的表现。据我们所知,这项工作代表了扩散模型在地标检测自监督学习中的首次应用,这可能为缓解数据稀缺的少量模式提供有价值的预训练方法。
论文及项目相关链接
PDF Accepted at WACV 2025
Summary
医学图像领域中深度神经网络的应用广泛,包括图像分类、分割和关键点检测等任务。然而,数据稀缺(包括可用的注释和图像)常常限制其应用。本研究首次将去噪扩散概率模型(DDPMs)应用于医学图像中的关键点检测任务,解决标注数据有限的问题。研究创新之处在于利用DDPMs进行自监督预训练,此方法在少量标注数据(仅50张图像)的情况下即可实现准确的关键点检测,并在三个流行的X射线基准数据集上超越了ImageNet监督预训练和传统自监督技术。
Key Takeaways
- 研究将去噪扩散概率模型(DDPMs)应用于医学图像中的关键点检测任务。
- DDPMs能有效解决医学图像中标注数据有限的问题。
- 研究采用自监督预训练方法,利用DDPMs在少量标注数据下实现准确的关键点检测。
- 该方法在三个流行的X射线基准数据集上表现优异,超越了传统的预训练技术。
- 此研究是首次将扩散模型应用于医学图像关键点检测中的自监督学习。
- 该方法为未来解决医学图像数据稀缺问题提供了新的思路。
点此查看论文截图

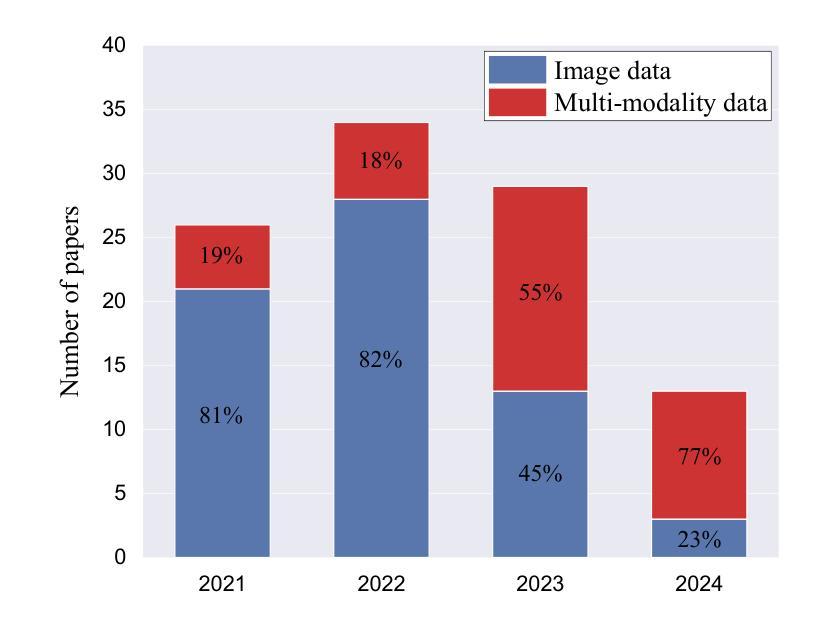
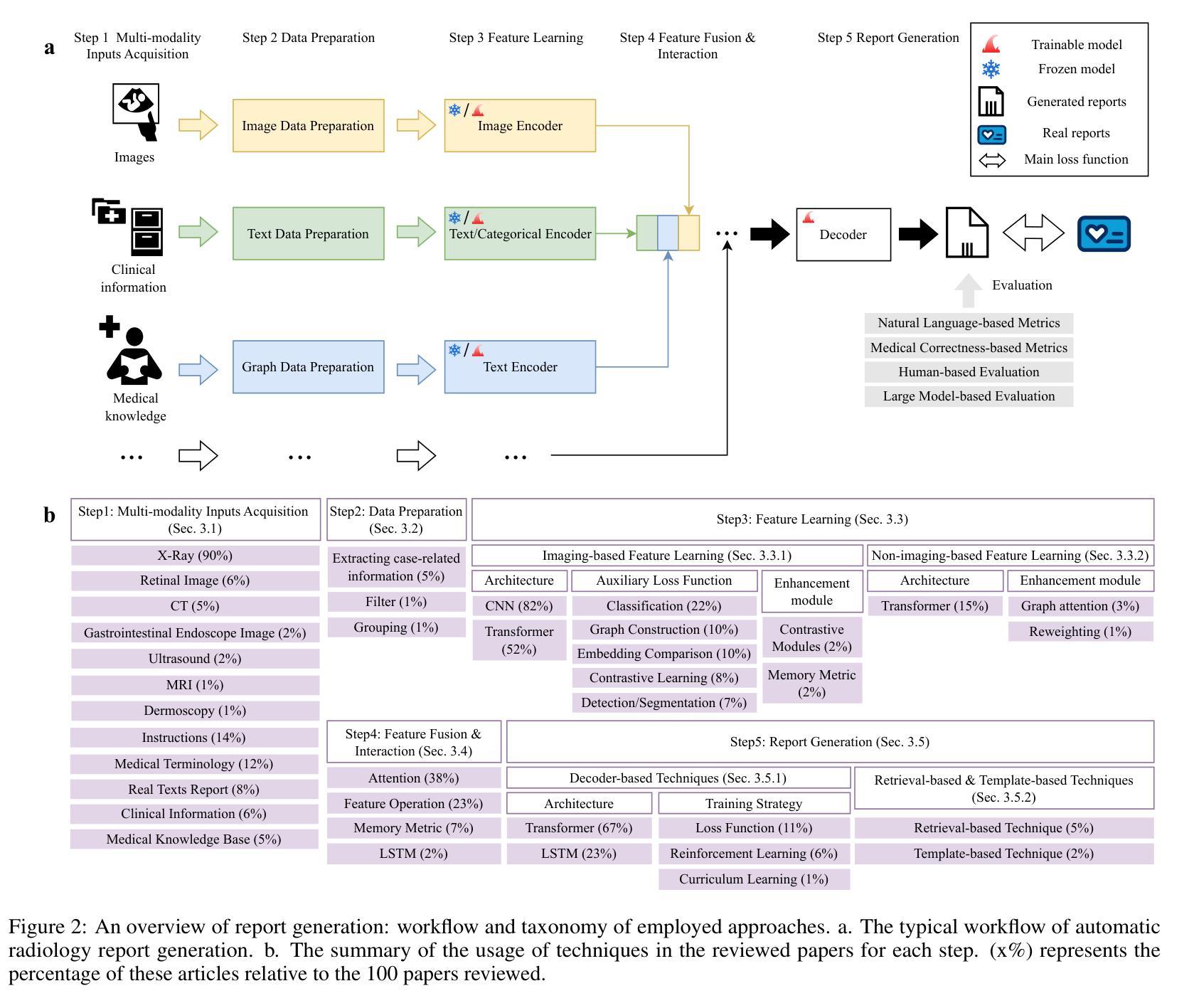
A Survey of Deep Learning-based Radiology Report Generation Using Multimodal Data
Authors:Xinyi Wang, Grazziela Figueredo, Ruizhe Li, Wei Emma Zhang, Weitong Chen, Xin Chen
Automatic radiology report generation can alleviate the workload for physicians and minimize regional disparities in medical resources, therefore becoming an important topic in the medical image analysis field. It is a challenging task, as the computational model needs to mimic physicians to obtain information from multi-modal input data (i.e., medical images, clinical information, medical knowledge, etc.), and produce comprehensive and accurate reports. Recently, numerous works have emerged to address this issue using deep-learning-based methods, such as transformers, contrastive learning, and knowledge-base construction. This survey summarizes the key techniques developed in the most recent works and proposes a general workflow for deep-learning-based report generation with five main components, including multi-modality data acquisition, data preparation, feature learning, feature fusion and interaction, and report generation. The state-of-the-art methods for each of these components are highlighted. Additionally, we summarize the latest developments in large model-based methods and model explainability, along with public datasets, evaluation methods, current challenges, and future directions in this field. We have also conducted a quantitative comparison between different methods in the same experimental setting. This is the most up-to-date survey that focuses on multi-modality inputs and data fusion for radiology report generation. The aim is to provide comprehensive and rich information for researchers interested in automatic clinical report generation and medical image analysis, especially when using multimodal inputs, and to assist them in developing new algorithms to advance the field.
自动放射学报告生成能够减轻医生的工作量,并最小化医疗资源的区域差异,因此成为医学图像分析领域的重要课题。这是一个具有挑战性的任务,因为计算模型需要模仿医生从多模态输入数据(例如医学图像、临床信息、医学知识等)中获得信息,并生成全面准确的报告。最近,出现了许多基于深度学习的方法来解决这个问题,例如变压器、对比学习和知识库构建。这篇综述总结了最近作品开发的关键技术,并提出了一个基于深度学习的报告生成的一般工作流程,包括五个主要组件:多模态数据采集、数据准备、特征学习、特征融合和交互以及报告生成。本文强调了这些组件的最新先进方法。此外,我们还总结了基于大型模型的最新发展和模型解释性,以及公开数据集、评估方法、当前挑战和该领域的未来发展方向。我们还在相同的实验环境中对不同方法进行了定量比较。这是最新专注于多模态输入和数据融合用于放射学报告生成的综述。旨在为对自动临床报告生成和医学图像分析感兴趣的研究人员提供全面丰富的信息,特别是在使用多模态输入时,并帮助他们开发新算法以推动该领域的发展。
论文及项目相关链接
Summary
本文介绍了自动放射学报告生成的重要性,包括其在减轻医生工作量、缩小医疗资源区域差距方面的作用。文章指出这是一个挑战任务,需要计算模型从多模态输入数据中获取信息并生成全面准确的报告。本文总结了最近利用深度学习的方法来解决这个问题,提出了基于深度学习报告的通用工作流程,并详细介绍了各部分的关键技术。同时进行了最新发展的大型模型方法、模型解释性等方面的总结。通过同一实验环境下不同方法的定量比较,本文是最新的关注多模态输入和数据融合的放射学报告生成的综述。旨在为对自动临床报告生成和医学图像分析感兴趣的学者提供全面丰富的信息,并帮助开发新算法推动该领域的发展。
Key Takeaways
- 自动放射学报告生成是医学图像分析领域的重要话题,有助于减轻医生工作量并缩小医疗资源差距。
- 该任务需要计算模型模仿医生从多模态输入数据中获取信息并生成准确全面的报告。
- 最近利用深度学习的方法来解决这个问题,包括transformer、对比学习和知识库构建等。
- 基于深度学习的报告生成包含五个主要组件:多模态数据采集、数据准备、特征学习、特征融合与交互和报告生成。
- 总结了各部分的关键技术和最新发展的大型模型方法、模型解释性等方面的内容。
- 文章通过定量比较不同方法在同一实验环境下的表现,提供了全面的综述。
点此查看论文截图

Implantable Adaptive Cells: A Novel Enhancement for Pre-Trained U-Nets in Medical Image Segmentation
Authors:Emil Benedykciuk, Marcin Denkowski, Grzegorz Wójcik
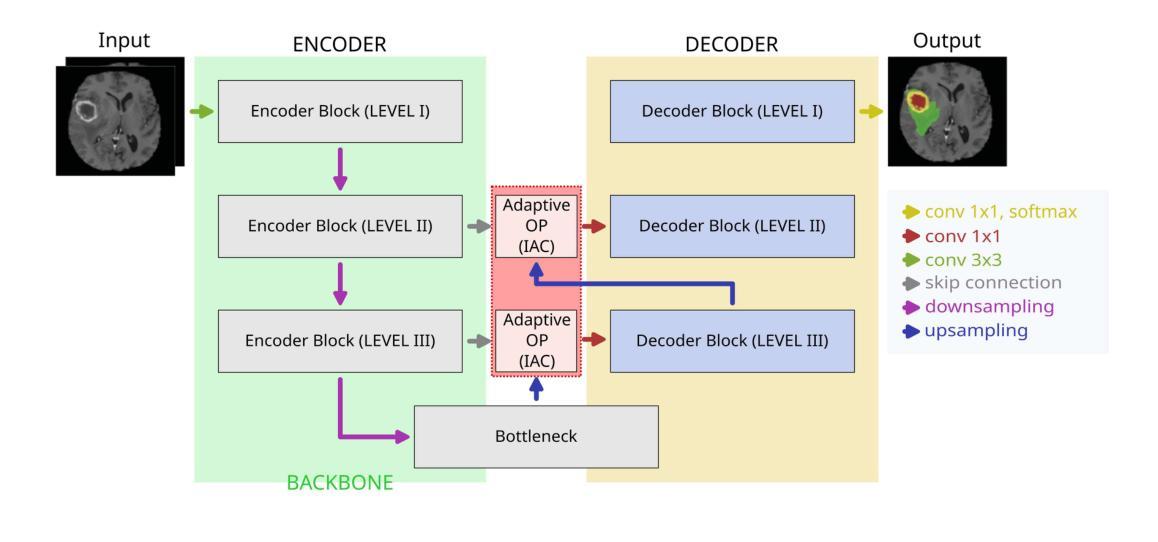
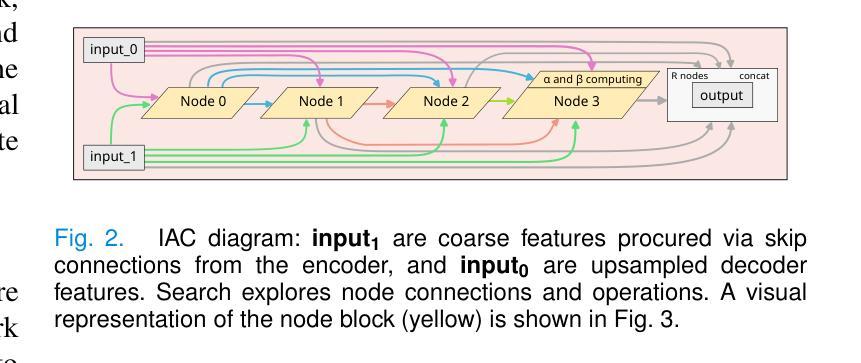
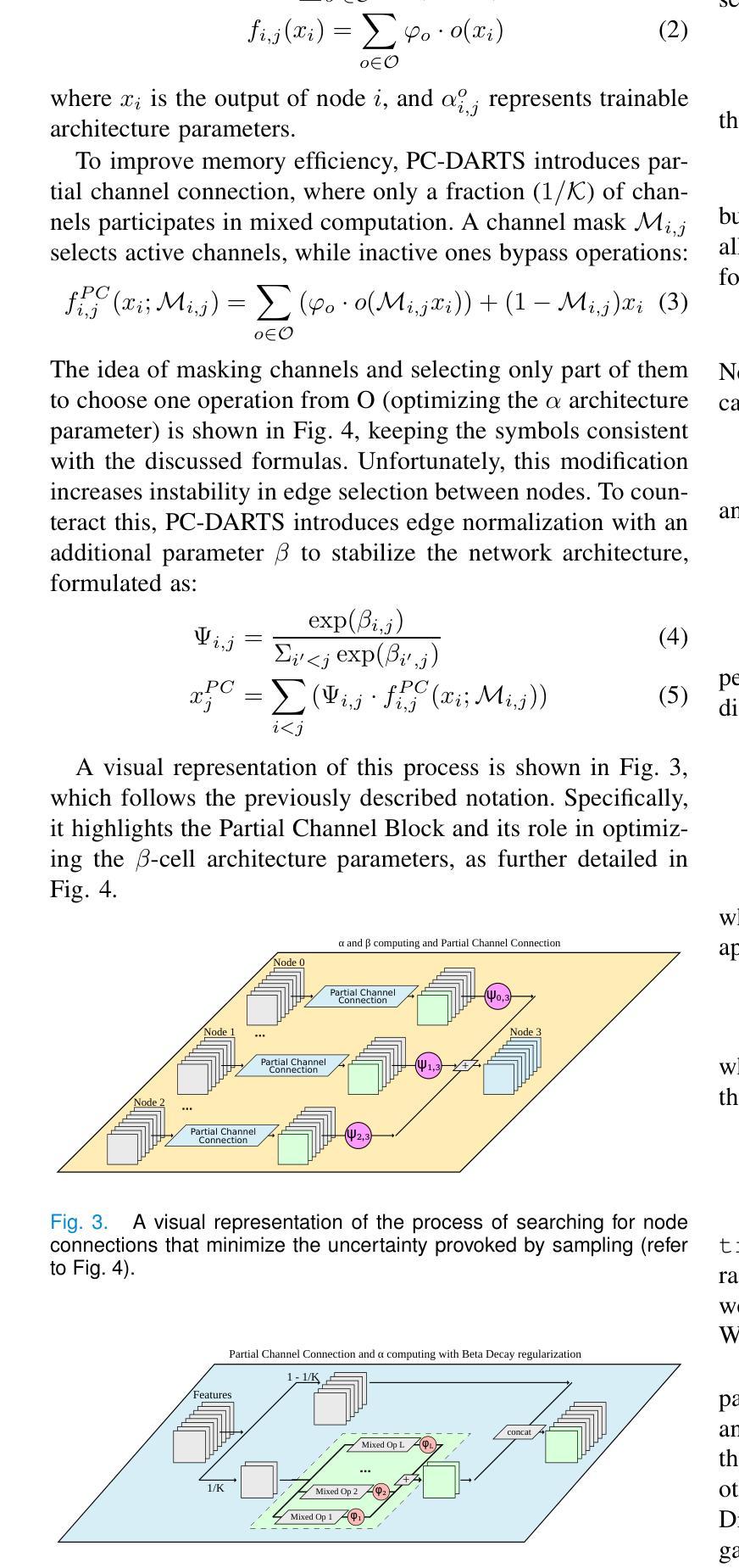
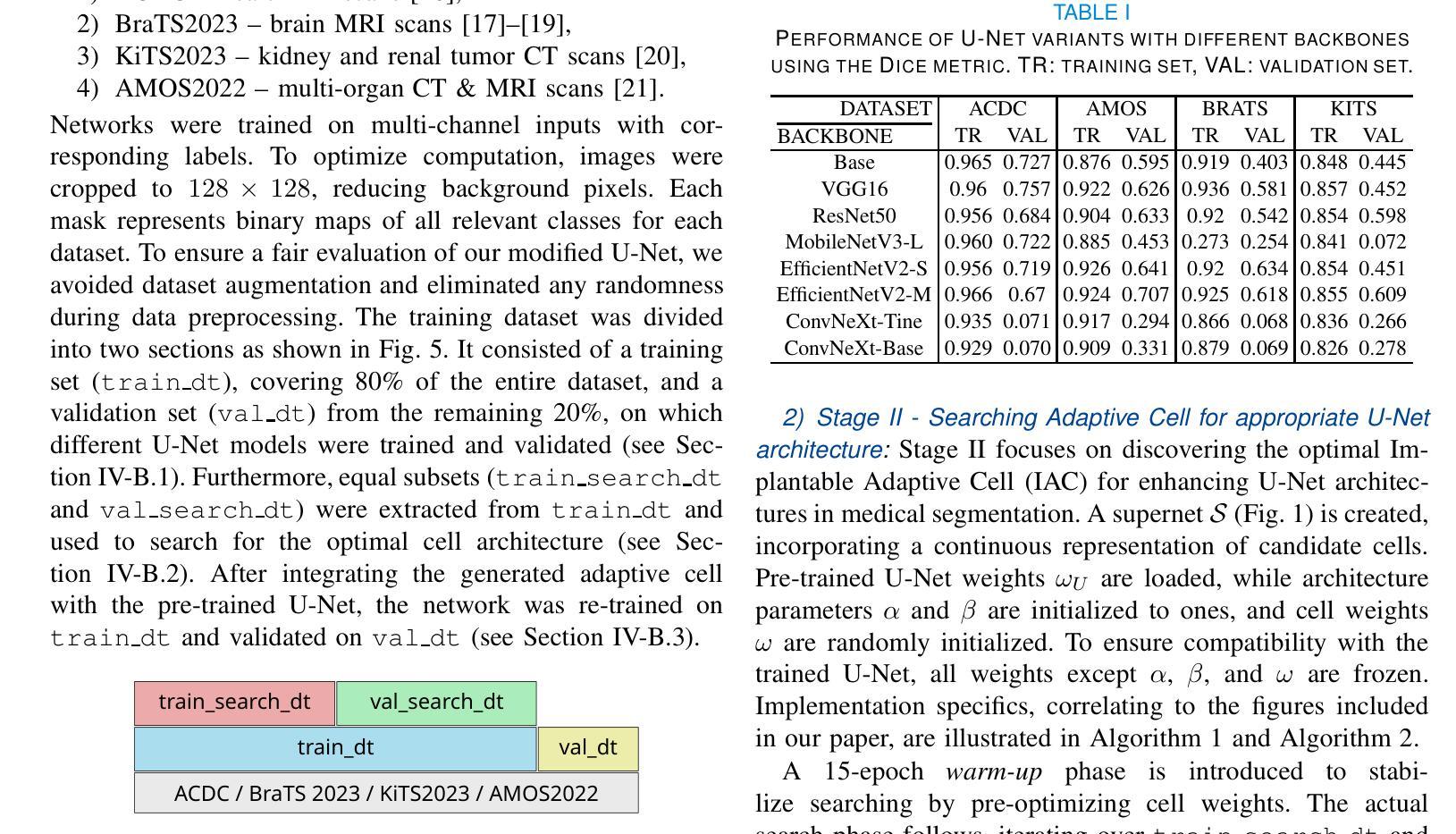
This paper introduces a novel approach to enhance the performance of pre-trained neural networks in medical image segmentation using gradient-based Neural Architecture Search (NAS) methods. We present the concept of Implantable Adaptive Cell (IAC), small modules identified through Partially-Connected DARTS based approach, designed to be injected into the skip connections of an existing and already trained U-shaped model. Unlike traditional NAS methods, our approach refines existing architectures without full retraining. Experiments on four medical datasets with MRI and CT images show consistent accuracy improvements on various U-Net configurations, with segmentation accuracy gain by approximately 5 percentage points across all validation datasets, with improvements reaching up to 11%pt in the best-performing cases. The findings of this study not only offer a cost-effective alternative to the complete overhaul of complex models for performance upgrades but also indicate the potential applicability of our method to other architectures and problem domains.
本文介绍了一种利用基于梯度的神经网络结构搜索(NAS)方法提高预训练神经网络在医学图像分割性能的新型方法。我们提出了可植入自适应单元(IAC)的概念,这是一种通过部分连接DARTS方法识别的小模块,旨在注入到已训练和存在的U形模型的跳过连接中。与传统的NAS方法不同,我们的方法能够在不进行全面再训练的情况下优化现有架构。在四个包含MRI和CT图像的医学数据集上进行的实验表明,在各种U-Net配置上,准确率得到持续提高,在所有验证数据集上的分割准确率提高约5个百分点,在表现最佳的案例中,提高幅度高达11个百分点。本研究的结果不仅为通过彻底改革复杂模型来提高性能提供了成本效益高的替代方案,还表明我们的方法在其他架构和问题领域具有潜在适用性。
论文及项目相关链接
Summary
论文提出了一种利用基于梯度的神经网络架构搜索(NAS)方法提高预训练神经网络在医学图像分割性能的新方法。通过部分连接的DARTS方法,设计了一种名为植入式自适应单元(IAC)的小模块,可注入已训练的U形模型的跳跃连接中。该方法在四个医学数据集上的实验表明,在各种U-Net配置上均实现了稳定的准确性提高,分割准确性平均提升约5个百分点,最佳情况下提升达11个百分点。此研究不仅为升级性能提供了全面推翻复杂模型的低成本替代方案,而且还表明该方法在其他架构和问题领域具有潜在适用性。
Key Takeaways
- 论文提出了一种基于梯度的新型神经网络架构搜索方法用于改进预训练神经网络在医学图像分割的性能。
- 采用名为植入式自适应单元(IAC)的小模块,通过部分连接的DARTS方法设计。
- IAC模块被注入到已训练的U形模型的跳跃连接中,以优化现有架构,无需完全重新训练。
- 在四个医学数据集上的实验表明,该方法在各种U-Net配置上都实现了分割准确性的显著提高。
- 与传统的NAS方法相比,该方法更为高效,成本更低。
- 该方法不仅适用于医学图像分割,还有潜力应用于其他架构和问题领域。
点此查看论文截图