⚠️ 以下所有内容总结都来自于 大语言模型的能力,如有错误,仅供参考,谨慎使用
🔴 请注意:千万不要用于严肃的学术场景,只能用于论文阅读前的初筛!
💗 如果您觉得我们的项目对您有帮助 ChatPaperFree ,还请您给我们一些鼓励!⭐️ HuggingFace免费体验
2025-03-11 更新
Task-oriented Uncertainty Collaborative Learning for Label-Efficient Brain Tumor Segmentation
Authors:Zhenxuan Zhang, Hongjie Wu, Jiahao Huang, Baihong Xie, Zhifan Gao, Junxian Du, Pete Lally, Guang Yang
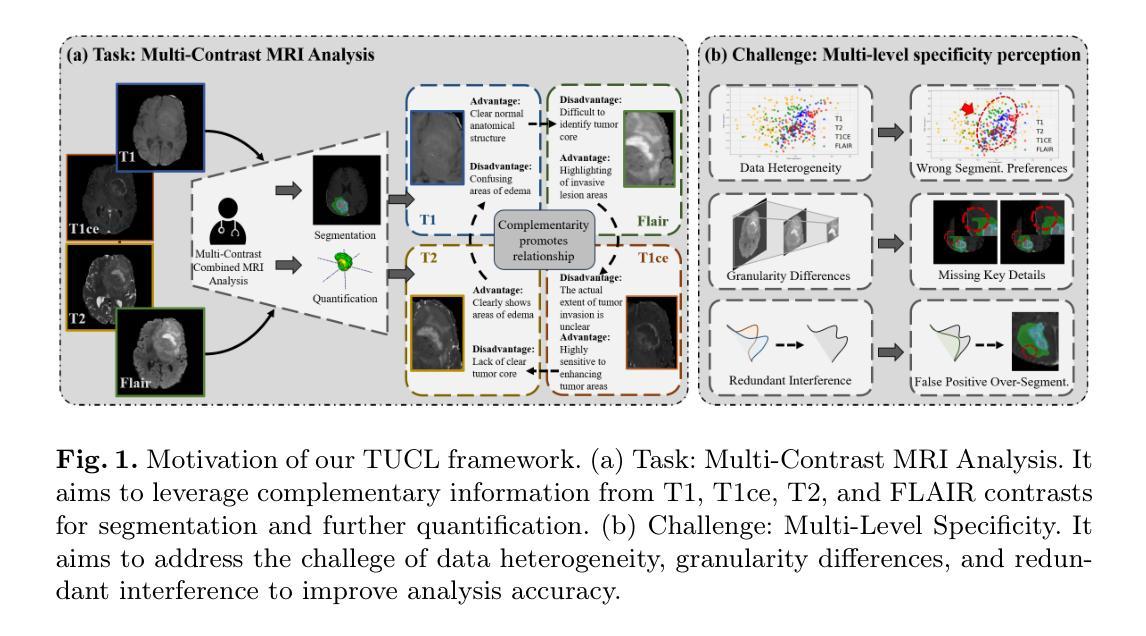
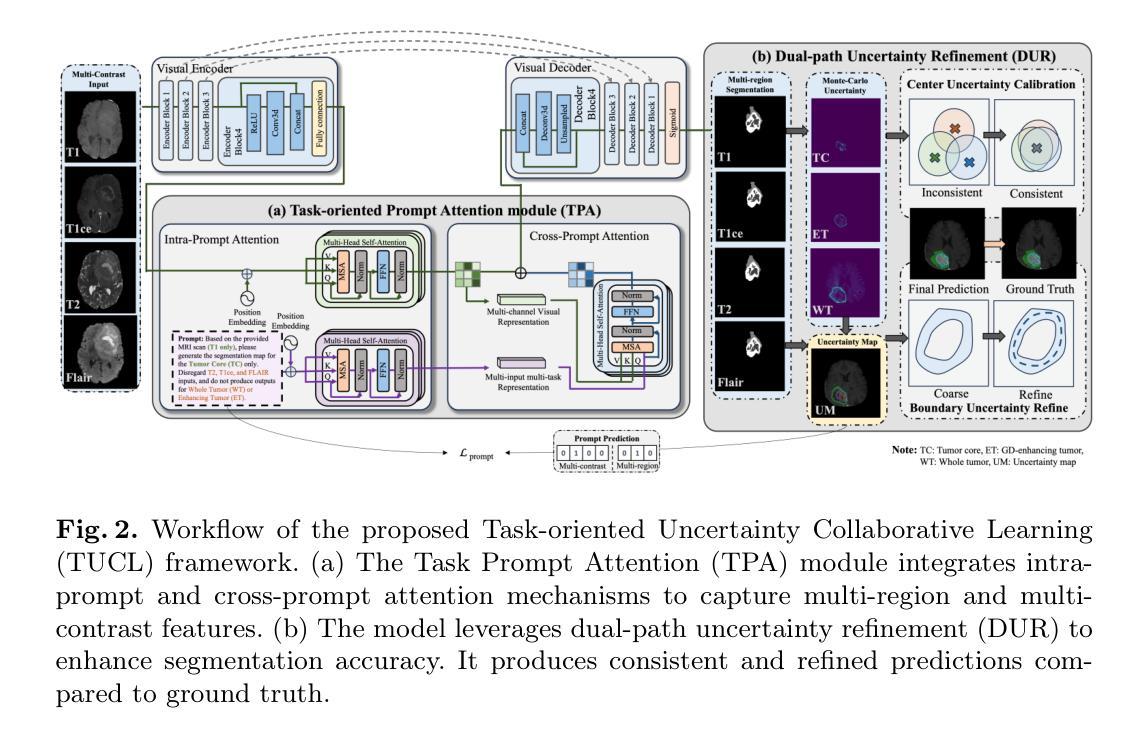
Multi-contrast magnetic resonance imaging (MRI) plays a vital role in brain tumor segmentation and diagnosis by leveraging complementary information from different contrasts. Each contrast highlights specific tumor characteristics, enabling a comprehensive understanding of tumor morphology, edema, and pathological heterogeneity. However, existing methods still face the challenges of multi-level specificity perception across different contrasts, especially with limited annotations. These challenges include data heterogeneity, granularity differences, and interference from redundant information. To address these limitations, we propose a Task-oriented Uncertainty Collaborative Learning (TUCL) framework for multi-contrast MRI segmentation. TUCL introduces a task-oriented prompt attention (TPA) module with intra-prompt and cross-prompt attention mechanisms to dynamically model feature interactions across contrasts and tasks. Additionally, a cyclic process is designed to map the predictions back to the prompt to ensure that the prompts are effectively utilized. In the decoding stage, the TUCL framework proposes a dual-path uncertainty refinement (DUR) strategy which ensures robust segmentation by refining predictions iteratively. Extensive experimental results on limited labeled data demonstrate that TUCL significantly improves segmentation accuracy (88.2% in Dice and 10.853 mm in HD95). It shows that TUCL has the potential to extract multi-contrast information and reduce the reliance on extensive annotations. The code is available at: https://github.com/Zhenxuan-Zhang/TUCL_BrainSeg.
多对比度磁共振成像(MRI)在脑肿瘤分割和诊断中扮演着重要角色,它通过利用来自不同对比度的补充信息来发挥作用。每个对比度都突出了特定的肿瘤特征,使我们能够全面了解肿瘤的形态、水肿和病理异质性。然而,现有方法仍然面临着不同对比度之间的多层次特异性感知挑战,尤其是标注数据有限的情况下。这些挑战包括数据异质性、粒度差异以及来自冗余信息的干扰。为了解决这些局限性,我们提出了面向任务的协同不确定性学习(TUCL)框架,用于多对比度MRI分割。TUCL引入了一个面向任务的提示注意力(TPA)模块,该模块具有内部提示和跨提示注意力机制,可以动态地建模跨对比度和任务的特征交互。此外,还设计了一个循环过程,将预测映射回提示,以确保有效地利用提示。在解码阶段,TUCL框架提出了一种双路径不确定性优化(DUR)策略,通过迭代优化预测,确保稳健的分割效果。在有限标注数据上的大量实验结果表明,TUCL显著提高了分割精度(Dice系数为88.2%,HD95为10.853mm)。这表明TUCL具有提取多对比度信息并减少对面标注依赖的潜力。代码可在:https://github.com/Zhenxuan-Zhang/TUCL_BrainSeg获取。
论文及项目相关链接
Summary
多对比磁共振成像(MRI)在脑肿瘤分割和诊断中扮演重要角色,通过不同对比信息揭示肿瘤特征。现有方法面临跨对比多层次特异性感知的挑战,如数据异质性、粒度差异和冗余信息干扰。为此,提出任务导向不确定性协同学习(TUCL)框架,包含任务导向提示注意力(TPA)模块和循环过程,实现跨对比和任务特征交互动态建模。解码阶段采用双路径不确定性优化(DUR)策略,确保在有限标注数据下精准分割。TUCL框架显著提高分割精度(Dice 88.2%,HD95为10.853mm),证明其挖掘多对比信息及减少对大量标注的依赖潜力。相关代码可访问:链接地址。
Key Takeaways
- 多对比磁共振成像(MRI)在脑肿瘤诊断与分割中作用关键,通过不同对比信息全面揭示肿瘤特性。
- 现有方法面临跨对比多层次特异性感知的挑战,包括数据异质性、粒度差异和冗余信息干扰。
- 提出的任务导向不确定性协同学习(TUCL)框架包括TPA模块和循环过程,实现特征交互的动态建模。
- TUCL框架采用双路径不确定性优化(DUR)策略,确保精准分割,即使在有限标注数据下。
点此查看论文截图

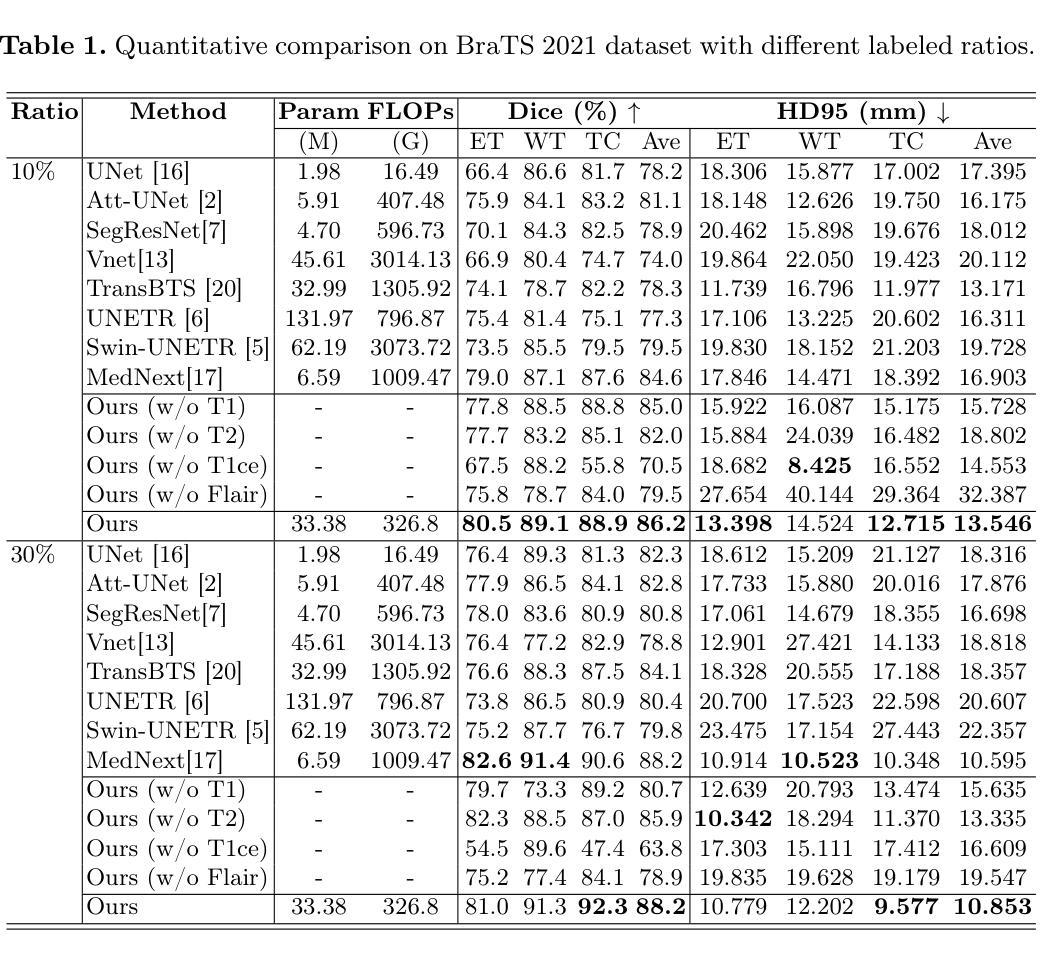
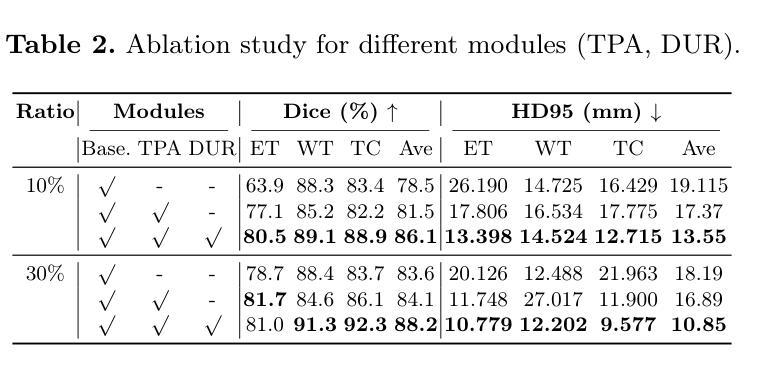
CACTUS: An Open Dataset and Framework for Automated Cardiac Assessment and Classification of Ultrasound Images Using Deep Transfer Learning
Authors:Hanae Elmekki, Ahmed Alagha, Hani Sami, Amanda Spilkin, Antonela Mariel Zanuttini, Ehsan Zakeri, Jamal Bentahar, Lyes Kadem, Wen-Fang Xie, Philippe Pibarot, Rabeb Mizouni, Hadi Otrok, Shakti Singh, Azzam Mourad
Cardiac ultrasound (US) scanning is a commonly used techniques in cardiology to diagnose the health of the heart and its proper functioning. Therefore, it is necessary to consider ways to automate these tasks and assist medical professionals in classifying and assessing cardiac US images. Machine learning (ML) techniques are regarded as a prominent solution due to their success in numerous applications aimed at enhancing the medical field, including addressing the shortage of echography technicians. However, the limited availability of medical data presents a significant barrier to applying ML in cardiology, particularly regarding US images of the heart. This paper addresses this challenge by introducing the first open graded dataset for Cardiac Assessment and ClassificaTion of UltraSound (CACTUS), which is available online. This dataset contains images obtained from scanning a CAE Blue Phantom and representing various heart views and different quality levels, exceeding the conventional cardiac views typically found in the literature. Additionally, the paper introduces a Deep Learning (DL) framework consisting of two main components. The first component classifies cardiac US images based on the heart view using a Convolutional Neural Network (CNN). The second component uses Transfer Learning (TL) to fine-tune the knowledge from the first component and create a model for grading and assessing cardiac images. The framework demonstrates high performance in both classification and grading, achieving up to 99.43% accuracy and as low as 0.3067 error, respectively. To showcase its robustness, the framework is further fine-tuned using new images representing additional cardiac views and compared to several other state-of-the-art architectures. The framework’s outcomes and performance in handling real-time scans were also assessed using a questionnaire answered by cardiac experts.
心脏超声(US)扫描是心脏病学中常用的技术,用于诊断心脏的健康状况和正常功能。因此,必须考虑自动化这些任务,并帮助医疗专业人士对心脏超声图像进行分类和评估。机器学习(ML)技术被视为一种突出的解决方案,因为它们在许多旨在改善医学领域的应用中取得了成功,包括解决超声技术人员短缺的问题。然而,医疗数据的有限可用性是心脏病学中应用机器学习的一个重大障碍,尤其是关于心脏的超声图像。本文通过引入心脏评估和分类超声(CACTUS)的第一个公开分级数据集来解决这一挑战,该数据集可在网上获得。该数据集包含从扫描CAE Blue Phantom获得的图像,代表不同的心脏视图和不同质量级别,超过了文献中通常发现的传统心脏视图。此外,本文介绍了一个深度学习(DL)框架,该框架包含两个主要组成部分。第一个组件使用卷积神经网络(CNN)根据心脏视图对心脏超声图像进行分类。第二个组件使用迁移学习(TL)对第一个组件的知识进行微调,并创建一个用于分级和评估心脏图像的模型。该框架在分类和分级方面都表现出出色的性能,分别达到了高达99.43%的准确率和低至0.3067的错误率。为了展示其稳健性,该框架使用代表其他心脏视图的图像进行了微调,并与其他一些最先进的架构进行了比较。框架处理实时扫描的结果和性能也通过心脏病专家回答的问卷进行了评估。
论文及项目相关链接
Summary
本文介绍了一种解决心脏超声扫描自动化分类和评估问题的方法。针对医学数据有限这一挑战,文章引入了首个公开分级的心脏超声图像评估与分类数据集CACTUS,包含从不同视角和不同质量水平扫描得到的图像。文章还提出了一种深度学习框架,包括基于卷积神经网络的心超图像视角分类和基于迁移学习的心脏图像分级评估模型。该框架在分类和分级方面表现出高性能,准确率高达99.43%,误差低至0.3067。并通过专家问卷评估了其在处理实时扫描中的稳健性和性能。
Key Takeaways
- 心脏超声扫描是诊断心脏健康和其功能的重要技术,自动化分类和评估这些图像对医疗专业人员的帮助很大。
- 机器学习技术在医疗领域有广泛应用,但医学数据的有限性是应用机器学习于心脏病学的一个重大挑战。
- 本文引入了CACTUS数据集,包含不同心脏视角和不同质量水平的图像,解决了医学数据有限的问题。
- 提出的深度学习框架包括基于CNN的心超图像视角分类和基于迁移学习的心脏图像分级评估模型。
- 框架在分类和分级方面表现出高性能,准确率高达99.43%,误差低。
- 该框架通过专家问卷评估了其在处理实时扫描中的稳健性和性能。
- 该研究为心脏超声扫描的自动化和智能化提供了新的思路和方法。
点此查看论文截图

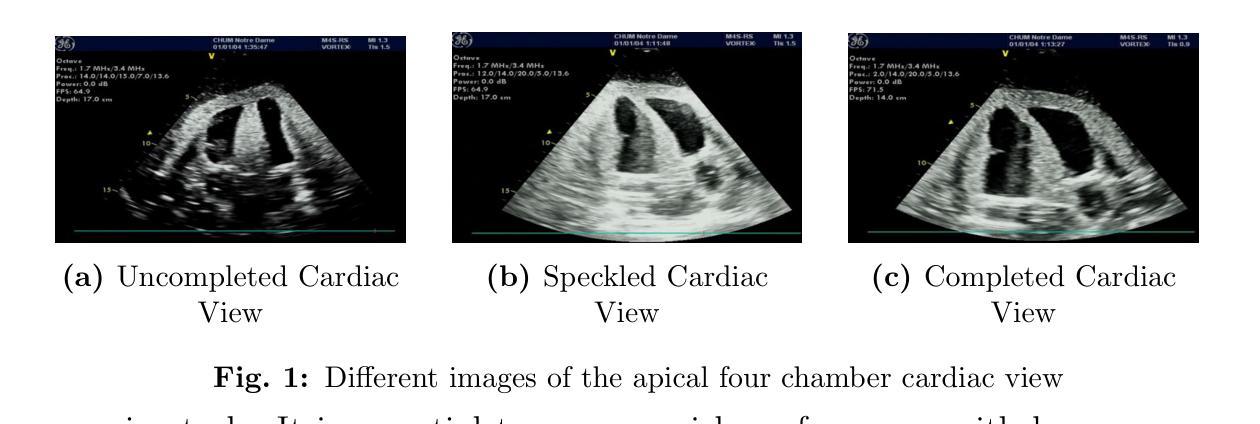
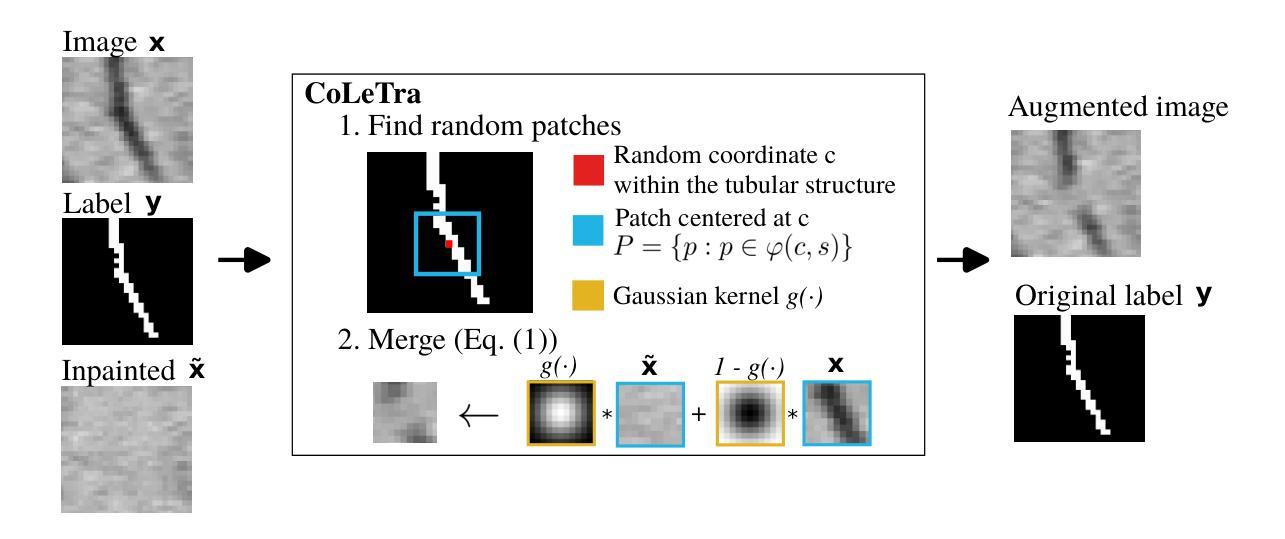
Disconnect to Connect: A Data Augmentation Method for Improving Topology Accuracy in Image Segmentation
Authors:Juan Miguel Valverde, Maja Østergaard, Adrian Rodriguez-Palomo, Peter Alling Strange Vibe, Nina Kølln Wittig, Henrik Birkedal, Anders Bjorholm Dahl
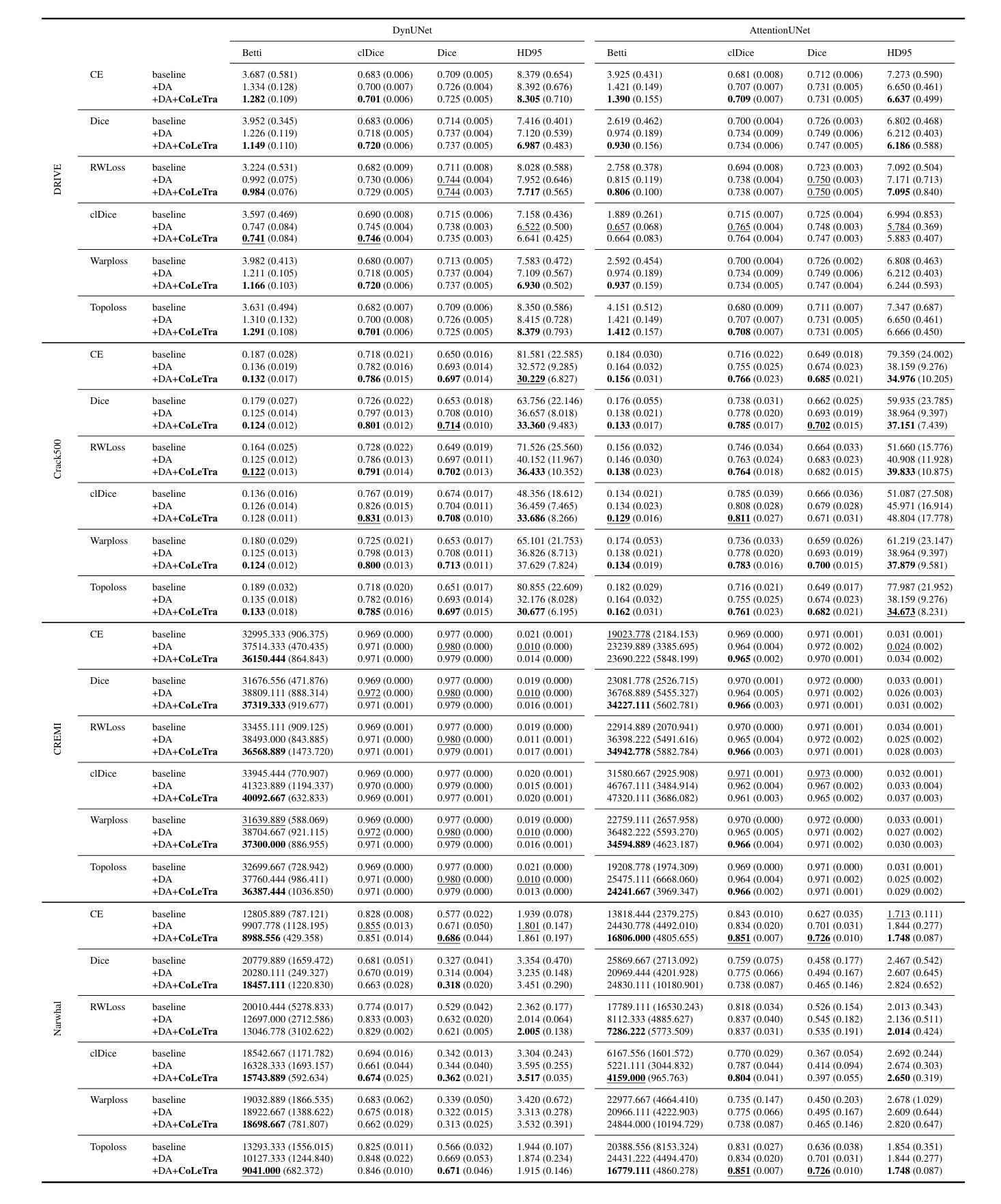
Accurate segmentation of thin, tubular structures (e.g., blood vessels) is challenging for deep neural networks. These networks classify individual pixels, and even minor misclassifications can break the thin connections within these structures. Existing methods for improving topology accuracy, such as topology loss functions, rely on very precise, topologically-accurate training labels, which are difficult to obtain. This is because annotating images, especially 3D images, is extremely laborious and time-consuming. Low image resolution and contrast further complicates the annotation by causing tubular structures to appear disconnected. We present CoLeTra, a data augmentation strategy that integrates to the models the prior knowledge that structures that appear broken are actually connected. This is achieved by creating images with the appearance of disconnected structures while maintaining the original labels. Our extensive experiments, involving different architectures, loss functions, and datasets, demonstrate that CoLeTra leads to segmentations topologically more accurate while often improving the Dice coefficient and Hausdorff distance. CoLeTra’s hyper-parameters are intuitive to tune, and our sensitivity analysis shows that CoLeTra is robust to changes in these hyper-parameters. We also release a dataset specifically suited for image segmentation methods with a focus on topology accuracy. CoLetra’s code can be found at https://github.com/jmlipman/CoLeTra.
精确分割薄管状结构(例如血管)对于深度神经网络来说是一项挑战。这些网络对单个像素进行分类,甚至轻微的误分类也可能会破坏这些结构中的薄连接。现有的提高拓扑精度的方法,如拓扑损失函数,依赖于非常精确且拓扑准确的训练标签,而这些标签的获取非常困难。这是因为标注图像,尤其是3D图像,是一项极其繁琐和耗时的任务。低图像分辨率和对比度会使管状结构出现断裂的外观,从而进一步使标注复杂化。我们提出了CoLeTra,这是一种数据增强策略,它将先验知识整合到模型中,即外观断裂的结构实际上是相连的。这是通过创建具有断裂结构外观的图像同时保持原始标签来实现的。我们进行了广泛的实验,涉及不同的架构、损失函数和数据集,证明CoLeTra在拓扑上实现了更精确的分割,同时经常提高Dice系数和Hausdorff距离。CoLeTra的超参数易于调整,我们的敏感性分析表明CoLeTra对这些超参数的变动具有稳健性。我们还发布了一个专门用于图像分割方法的数据集,重点关注拓扑精度。CoLeTra的代码可在https://github.com/jmlipman/CoLeTra找到。
论文及项目相关链接
Summary
本文提出一种名为CoLeTra的数据增强策略,用于改进深度学习模型对薄管状结构(如血管)的分割准确性。该策略通过生成看似断裂的结构图像同时保留原始标签,使模型学习到实际连接的先验知识。实验表明,CoLeTra能提高拓扑准确性,同时改善Dice系数和Hausdorff距离。
Key Takeaways
- 深度学习在分割薄管状结构(如血管)时面临挑战,因微小误分类可能导致结构断裂。
- 现有提高拓扑准确性的方法依赖精确的训练标签,但获取这些标签极为耗时。
- CoLeTra是一种数据增强策略,使模型学习到断裂结构实际连接的先验知识。
- CoLeTra通过生成看似断裂的结构图像同时保留原始标签来实现这一目的。
- 实验表明,CoLeTra能提高拓扑准确性,同时改善评估分割质量的Dice系数和Hausdorff距离。
- CoLeTra的超参数易于调整,且对参数变化具有鲁棒性。
点此查看论文截图

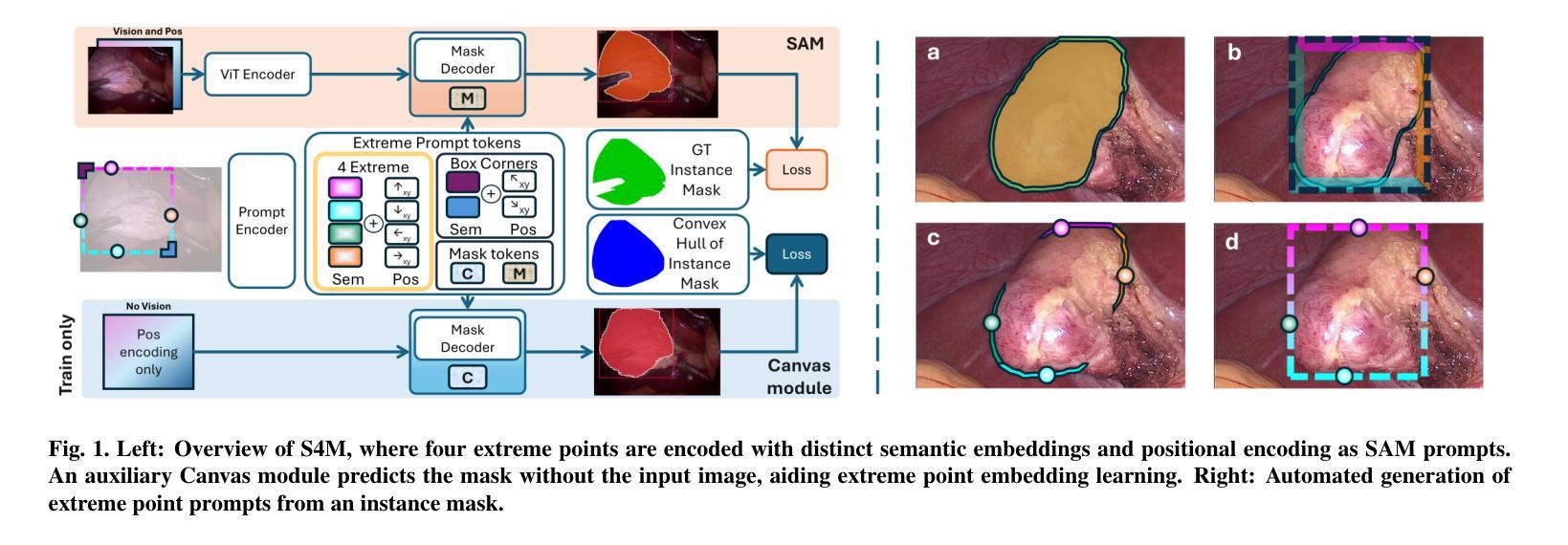
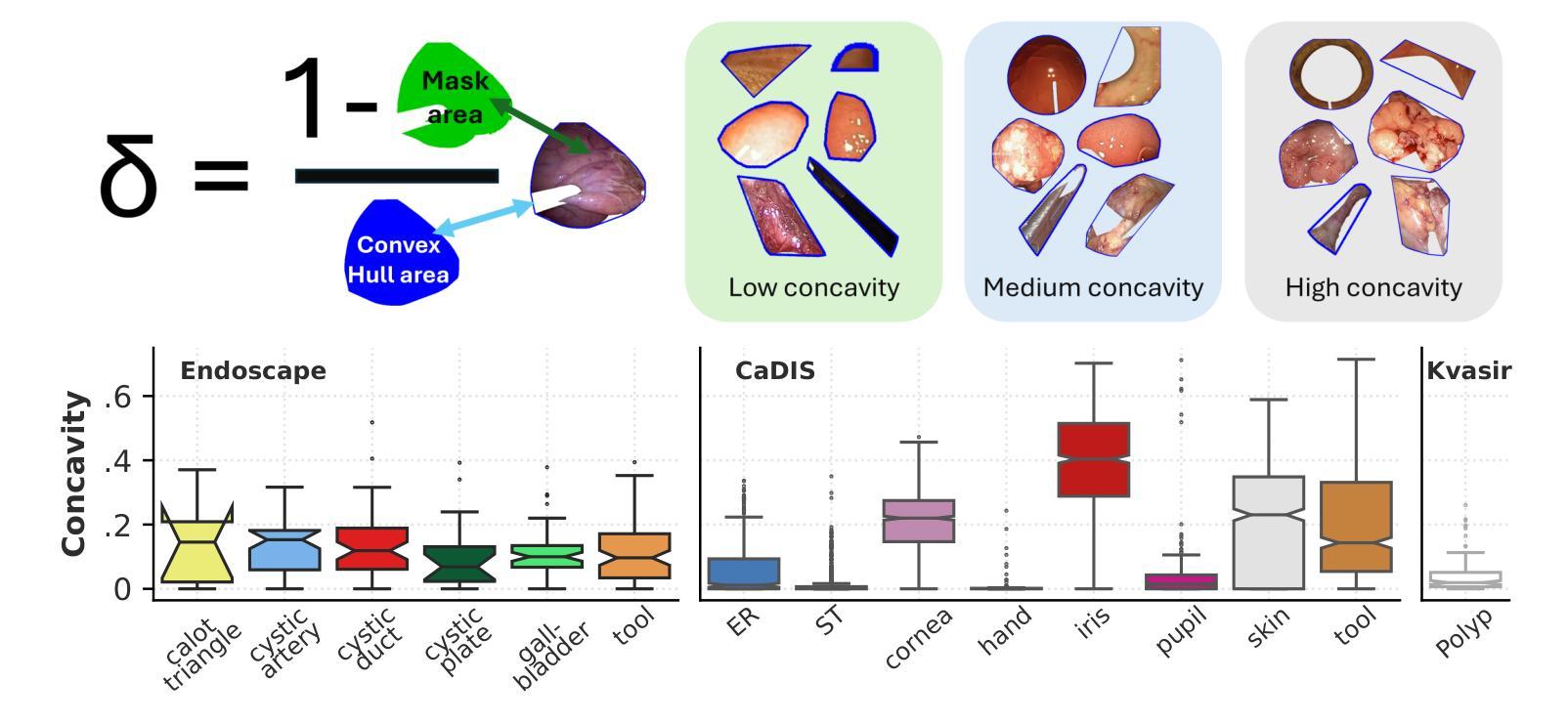
S4M: Segment Anything with 4 Extreme Points
Authors:Adrien Meyer, Lorenzo Arboit, Giuseppe Massimiani, Francesco Brucchi, Luca Emanuele Amodio, Didier Mutter, Nicolas Padoy
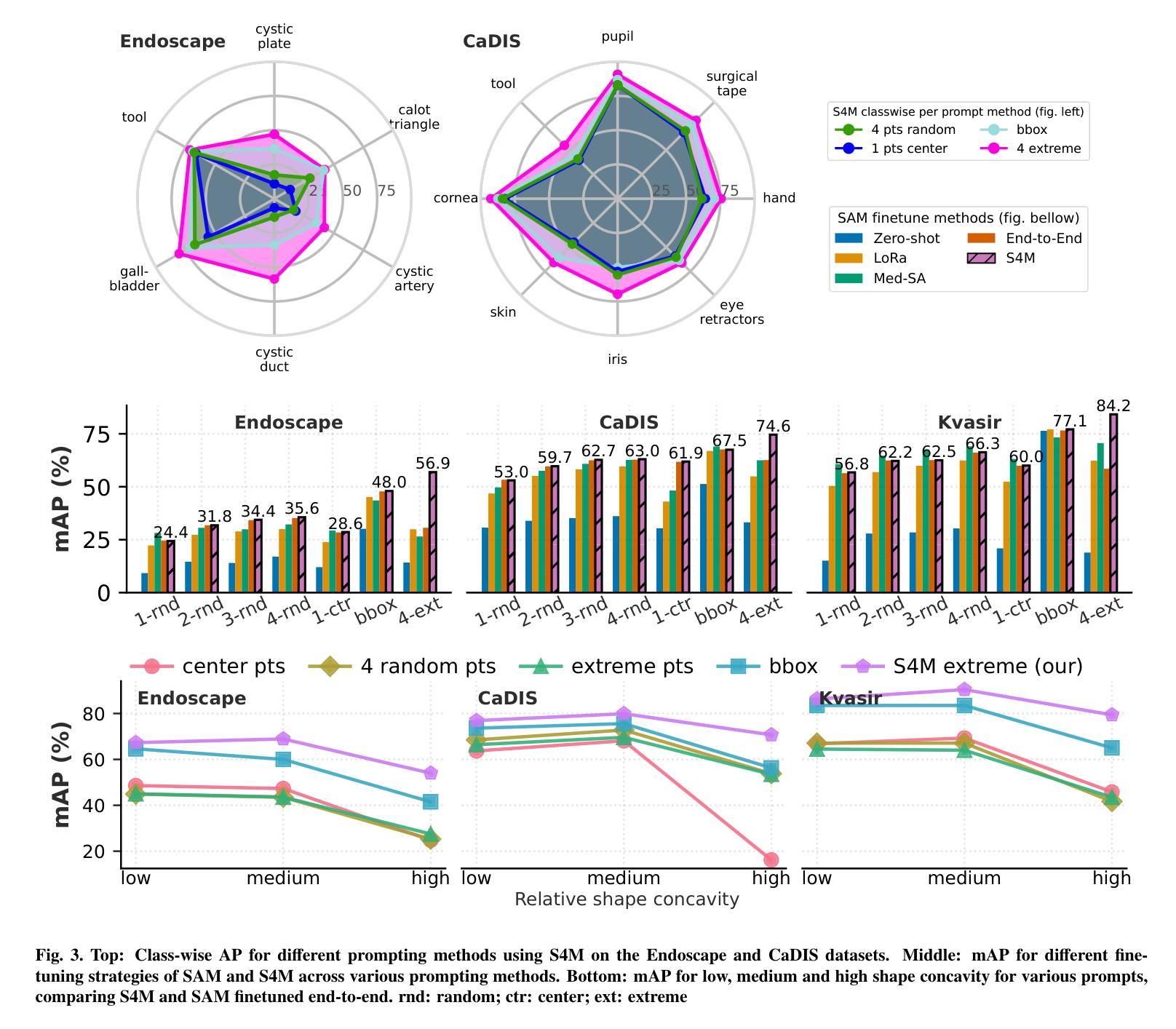
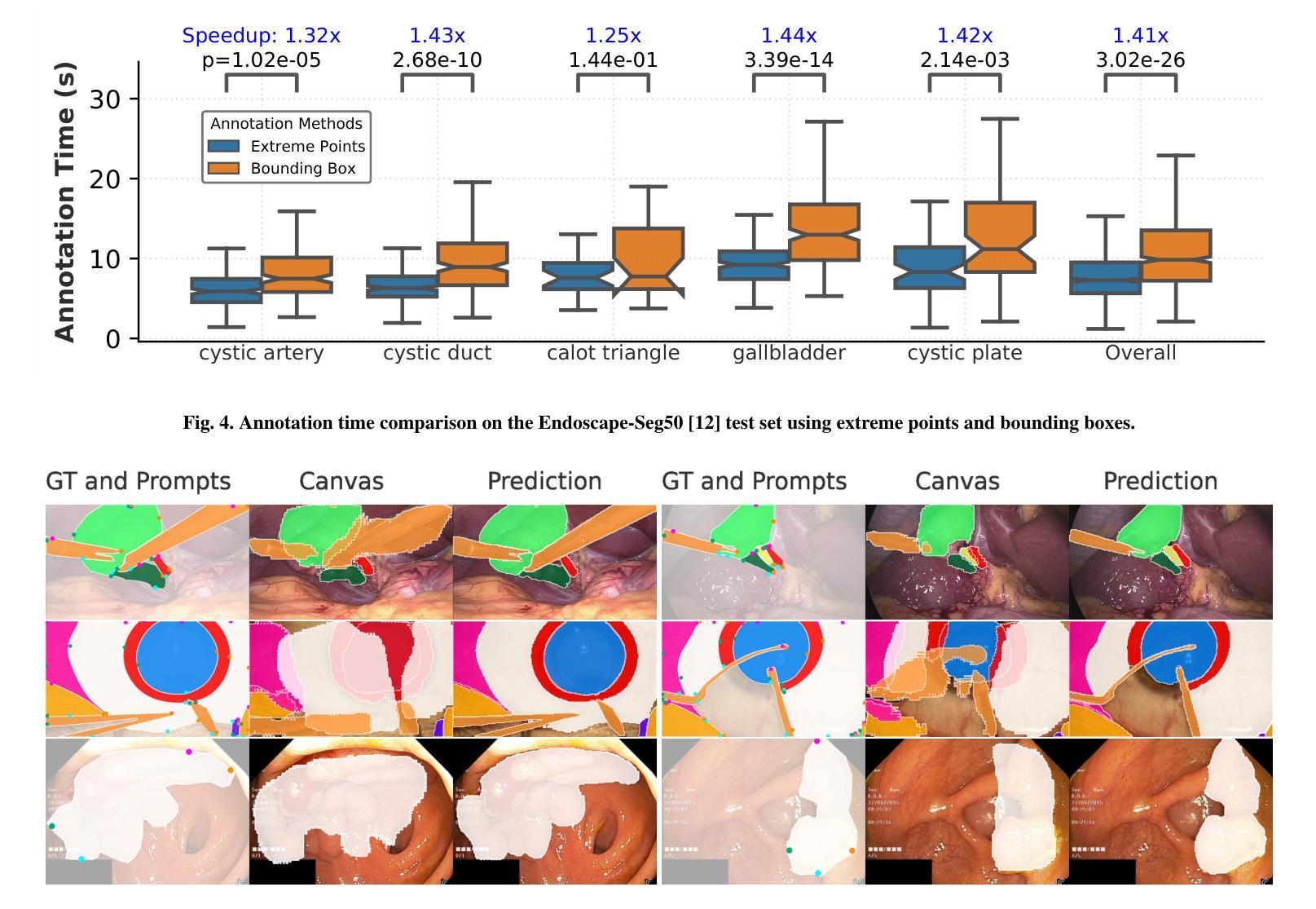
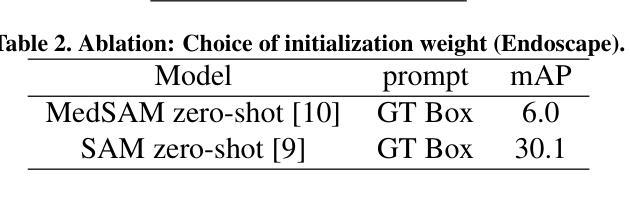
The Segment Anything Model (SAM) has revolutionized open-set interactive image segmentation, inspiring numerous adapters for the medical domain. However, SAM primarily relies on sparse prompts such as point or bounding box, which may be suboptimal for fine-grained instance segmentation, particularly in endoscopic imagery, where precise localization is critical and existing prompts struggle to capture object boundaries effectively. To address this, we introduce S4M (Segment Anything with 4 Extreme Points), which augments SAM by leveraging extreme points – the top-, bottom-, left-, and right-most points of an instance – prompts. These points are intuitive to identify and provide a faster, structured alternative to box prompts. However, a na"ive use of extreme points degrades performance, due to SAM’s inability to interpret their semantic roles. To resolve this, we introduce dedicated learnable embeddings, enabling the model to distinguish extreme points from generic free-form points and better reason about their spatial relationships. We further propose an auxiliary training task through the Canvas module, which operates solely on prompts – without vision input – to predict a coarse instance mask. This encourages the model to internalize the relationship between extreme points and mask distributions, leading to more robust segmentation. S4M outperforms other SAM-based approaches on three endoscopic surgical datasets, demonstrating its effectiveness in complex scenarios. Finally, we validate our approach through a human annotation study on surgical endoscopic videos, confirming that extreme points are faster to acquire than bounding boxes.
Segment Anything Model(SAM)已经彻底改变了开放集交互式图像分割,并为医学领域激发了众多适配器。然而,SAM主要依赖于稀疏提示,如点或边界框,这可能对于精细粒度的实例分割来说并不理想,特别是在内窥镜影像中,精确的定位至关重要,而现有的提示很难有效地捕捉对象边界。为了解决这一问题,我们推出了S4M(使用四个极点进行任何分割),它通过利用实例的极点——最顶部、底部、左侧和右侧的点——来增强SAM的提示。这些点易于识别,并提供了一种更快、更结构化的替代框提示。然而,极端点的直接使用会降低性能,因为SAM无法解释它们的语义角色。为了解决这一问题,我们引入了专用的可学习嵌入,使模型能够区分极端点和一般的自由形式点,并更好地推理它们之间的空间关系。我们进一步提出了一个辅助训练任务,通过Canvas模块,该模块仅通过提示进行操作——没有视觉输入——来预测粗略的实例掩码。这鼓励模型内化极点和掩码分布之间的关系,从而实现更稳健的分割。S4M在三个内窥镜手术数据集上的表现优于其他SAM方法,证明了其在复杂场景中的有效性。最后,我们通过关于手术内窥镜视频的人类注释研究验证了我们的方法,确认极点比边界框更快获取。
论文及项目相关链接
Summary
SAM模型在开放集交互式图像分割领域具有革命性影响,并为医学领域提供了许多适配器。然而,SAM主要依赖于稀疏提示,如点或边界框,对于精细的实例分割,特别是在内窥镜影像中,可能不够理想。为此,我们引入了S4M(通过四点进行任何分割),它通过利用实例的最上、最下、最左和最右四点提示来增强SAM。这些点易于识别,提供了更快的结构化替代方案。然而,单纯使用极点会降低性能,因为SAM无法解释它们的语义角色。为解决这一问题,我们引入了专用可学习嵌入,使模型能够区分极点和通用自由形式点,并更好地推理它们之间的空间关系。我们的方法在三个内窥镜手术数据集上优于其他SAM方法,验证了其在复杂场景中的有效性。最终,我们通过关于手术内窥镜视频的人类注释研究验证了我们的方法,确认极点比边界框更容易获取。
Key Takeaways
- SAM模型在开放集交互式图像分割中的重要作用,并描述了其对医学领域的影响。
- SAM主要依赖于稀疏提示(如点和边界框)进行图像分割,这在精细的实例分割中可能存在问题。
- S4M通过利用实例的四个极点(最上、最下、最左、最右)来增强SAM,这些点提供了一种更快速和结构化的替代方案。
- 单纯使用极点会降低性能,因为SAM无法解释它们的语义角色。为解决此问题,引入了专用可学习嵌入和辅助训练任务。
- S4M在三个内窥镜手术数据集上的表现优于其他SAM方法,显示了其在复杂场景中的有效性。
点此查看论文截图

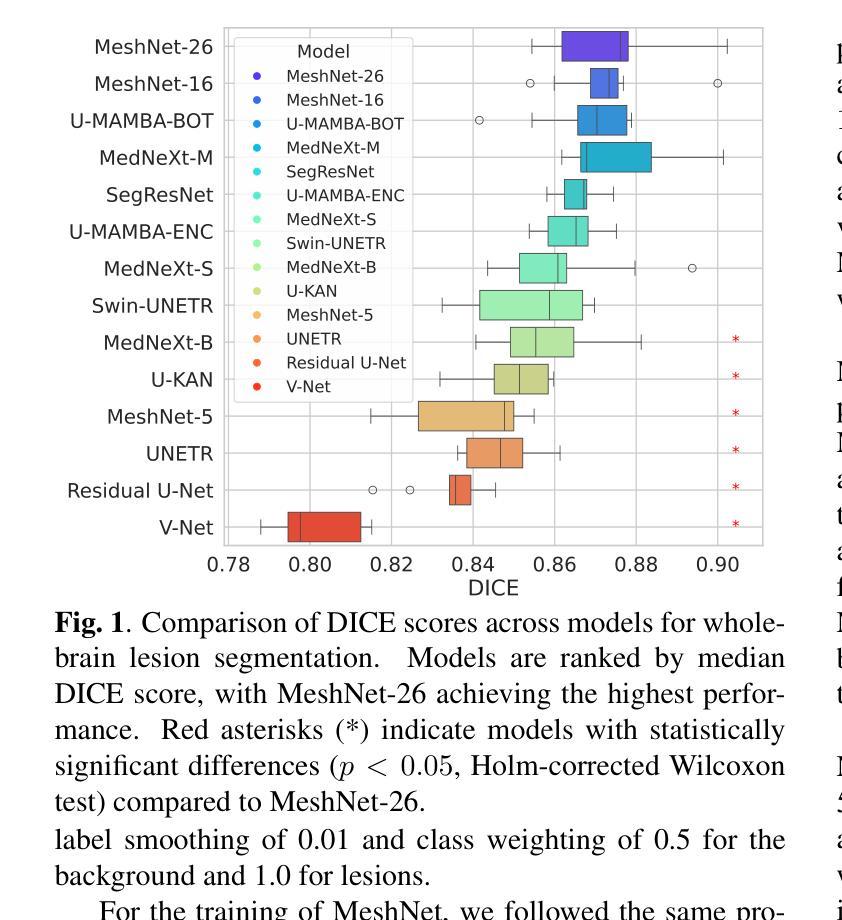
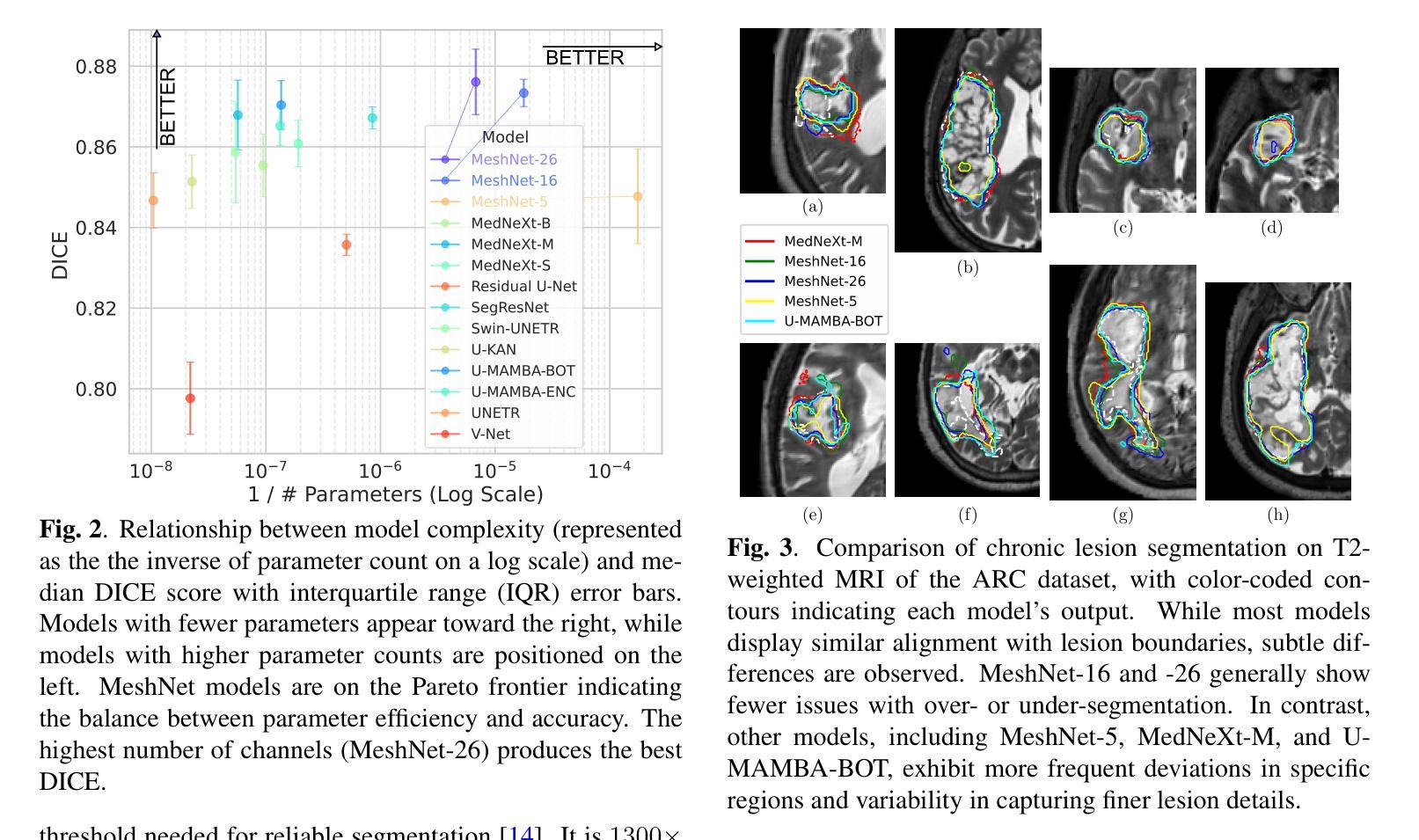
State-of-the-Art Stroke Lesion Segmentation at 1/1000th of Parameters
Authors:Alex Fedorov, Yutong Bu, Xiao Hu, Chris Rorden, Sergey Plis
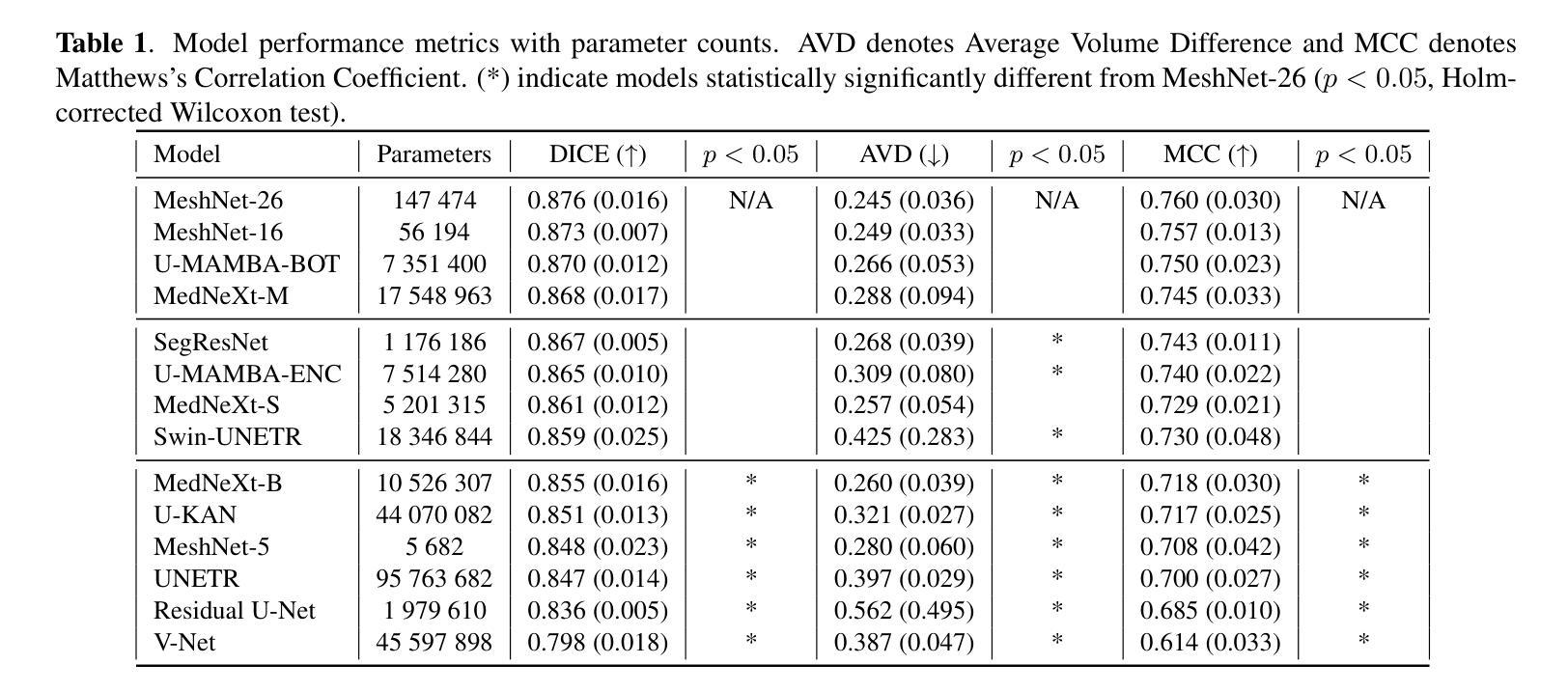
Efficient and accurate whole-brain lesion segmentation remains a challenge in medical image analysis. In this work, we revisit MeshNet, a parameter-efficient segmentation model, and introduce a novel multi-scale dilation pattern with an encoder-decoder structure. This innovation enables capturing broad contextual information and fine-grained details without traditional downsampling, upsampling, or skip-connections. Unlike previous approaches processing subvolumes or slices, we operate directly on whole-brain $256^3$ MRI volumes. Evaluations on the Aphasia Recovery Cohort (ARC) dataset demonstrate that MeshNet achieves superior or comparable DICE scores to state-of-the-art architectures such as MedNeXt and U-MAMBA at 1/1000th of parameters. Our results validate MeshNet’s strong balance of efficiency and performance, making it particularly suitable for resource-limited environments such as web-based applications and opening new possibilities for the widespread deployment of advanced medical image analysis tools.
在医学图像分析中,高效且准确的全脑病变分割仍然是一个挑战。在这项工作中,我们重新研究了MeshNet(一种参数高效的分割模型),并引入了一种新的多尺度膨胀模式,该模式具有编码器-解码器结构。这种创新使得能够在不采用传统下采样、上采样或跳过连接的情况下捕获广泛的上下文信息和精细细节。与以往处理子体积或切片的方法不同,我们直接在全脑的$ 256^3 $ MRI体积上进行操作。在阿弗西亚恢复队列(ARC)数据集上的评估表明,MeshNet的DICE得分与最新架构MedNeXt和U-MAMBA相比表现优越或相当,而其参数仅为后者的千分之一。我们的结果验证了MeshNet在效率和性能方面的强大平衡性,使其特别适用于资源受限的环境,如基于Web的应用程序,并为高级医学图像分析工具的广泛部署开辟了新的可能性。
论文及项目相关链接
PDF International Symposium on Biomedical Imaging, April 14-17, 2025
Summary
高效且精准的全脑病变分割在医学图像分析中仍是一个挑战。本研究重新考察了MeshNet这一参数高效的分割模型,并引入了一种新的多尺度膨胀模式与编码器-解码器结构。此创新能在不采用传统下采样、上采样或跳过连接的情况下,捕捉广泛上下文信息和精细细节。不同于处理子体积或切片的先前方法,我们直接在整脑$256^3$的MRI体积上进行操作。在阿弗西亚恢复队列(ARC)数据集上的评估表明,MeshNet的DICE得分与最新架构MedNeXt和U-MAMBA相比具有优越性或可比性,而所需的参数只有后者的千分之一。我们的结果验证了MeshNet在效率和性能之间的出色平衡,使其成为资源受限环境(如基于网络的应用程序)的理想选择,并为先进的医学图像分析工具的广泛部署提供了新的可能性。
Key Takeaways
- MeshNet模型在医学图像分析中具有高效和准确的分割能力。
- 引入新的多尺度膨胀模式与编码器-解码器结构,捕捉广泛上下文信息和精细细节。
- 直接在整脑MRI体积上进行操作,无需传统下采样、上采样或跳过连接。
- 在ARC数据集上的评估显示,MeshNet的DICE得分与最新架构相比具有优越性或相当。
- MeshNet模型参数效率极高,仅使用最新架构千分之一的参数。
- MeshNet在效率和性能之间达到良好平衡,适合资源受限环境。
点此查看论文截图

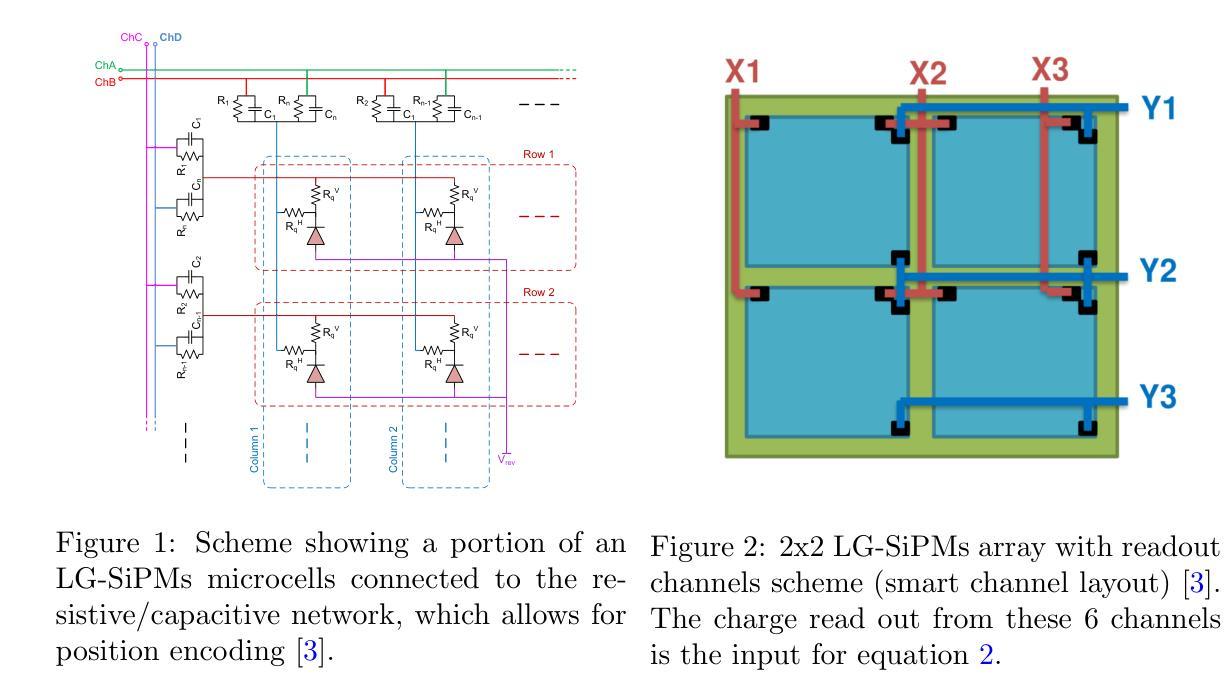
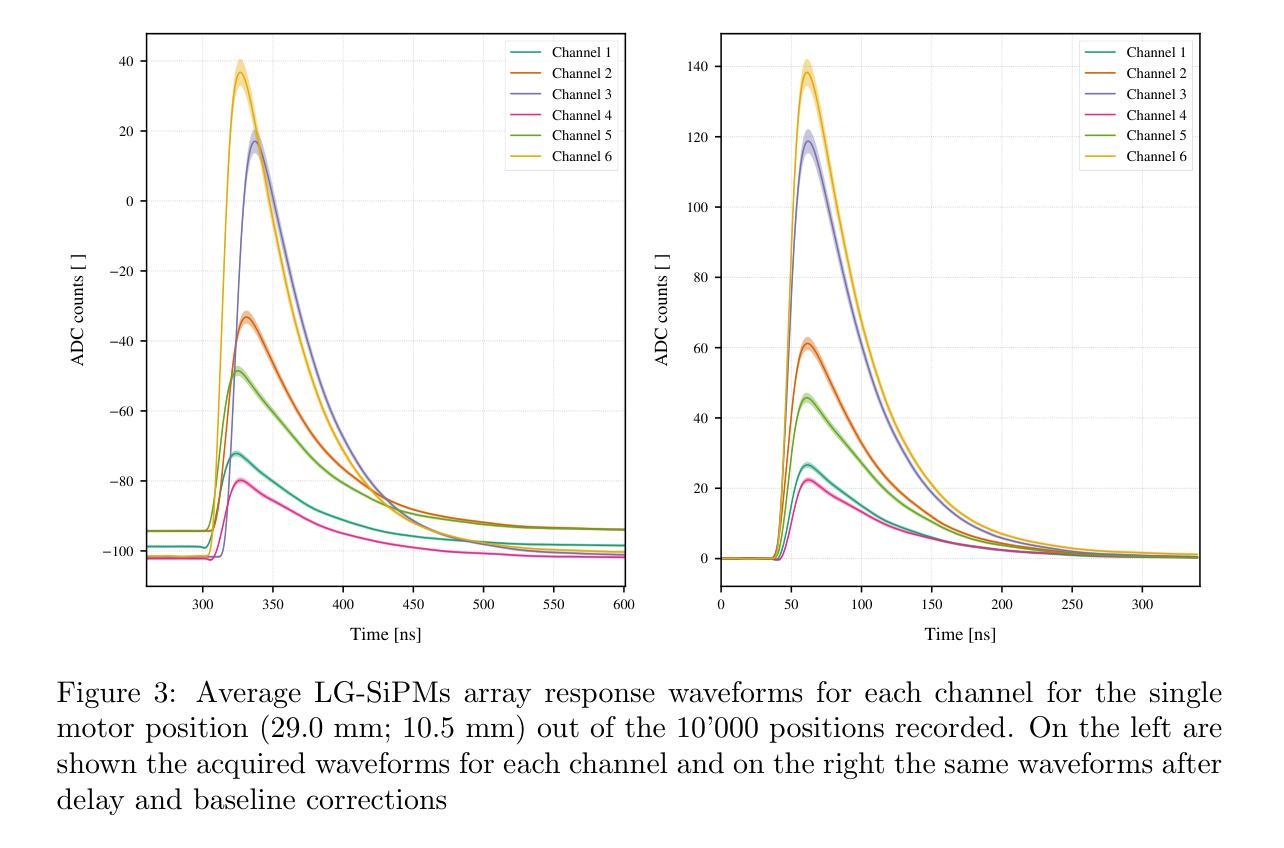
Quantitative Determination of Spatial Resolution and Linearity of Position-Sensitive LG-SiPMs at Sub-Millimeter Scale via Ricean Distribution Fitting
Authors:Aramis Raiola, Fabio Acerbi, Cyril Alispach, Hossein Arabi, Domenico della Volpe, Alberto Gola, Habib Zaidi
Position-sensitive SiPMs are useful in all light detection applications requiring a small number of readout channels while preserving the information about the incoming light’s interaction position. Focusing on a 2x2 array of LG-SiPMs covering an area of $\sim 15.5 \times 15.5\rm{mm}$ with just 6 readout channels, we proposed a quantitative method to evaluate image reconstruction performance. The method is based on a statistical approach to assess the device’s precision (spatial resolution) and accuracy (linearity) in reconstructing the light spot center of gravity. This evaluation is achieved through a Rice probability distribution function fitting. We obtained an average sensor spatial resolution’s best value of $81 \pm 3\rm{\mu m}$ (standard deviation), which is achieved by reconstructing each position with the amplitude of the channels’ output signals. The corresponding accuracy is $231 \pm 4~\rm{\mu m}$.
位置敏感的SiPM在所有需要少量读出通道同时保留关于入射光交互位置信息的光检测应用中都有用。我们重点关注一个由LG-SiPM组成的2x2阵列,覆盖约$ 15.5 \times 15.5毫米$的区域,仅使用6个读出通道。我们提出了一种定量评估图像重建性能的方法。该方法基于统计方法评估设备在重建光斑重心时的精度(空间分辨率)和准确度(线性度)。这种评估是通过拟合Rice概率分布函数来实现的。我们获得了传感器平均空间分辨率的最佳值,为$ 81 \pm 3微米$(标准偏差),这是通过根据各通道的输出来重建每个位置而实现的。相应的准确度为$ 231 \pm 4微米$。
论文及项目相关链接
PDF 17 pages, 9 figures, 2 tables
Summary
位置敏感的SiPM在需要少量读出通道的同时保留关于入射光交互位置的信息,适用于所有光检测应用。针对一个覆盖约$ 15.5 \times 15.5 \rm{mm}$区域,仅有6个读出通道的LG-SiPM的2x2阵列,提出了一种定量评估图像重建性能的方法。该方法基于统计方法评估设备的精度(空间分辨率)和准确性(线性度),通过拟合Rice概率分布函数实现光斑重心重建的评估。最佳传感器空间分辨率为$ 81 \pm 3 \rm{\mu m}$(标准偏差)。
Key Takeaways
- 位置敏感的SiPM适用于光检测应用,尤其是需要保留光交互位置信息的情况。
- 对于LG-SiPM的2x2阵列,即使使用有限的读出通道,也能实现图像重建性能的评价。
- 采用统计方法评估设备的空间分辨率和准确性。
- 通过拟合Rice概率分布函数来评估光斑重心的重建效果。
- 最佳传感器空间分辨率达到$ 81 \pm 3 \rm{\mu m}$。
- 设备在光斑重建方面的准确性为$ 231 \pm 4 \rm{\mu m}$。
点此查看论文截图

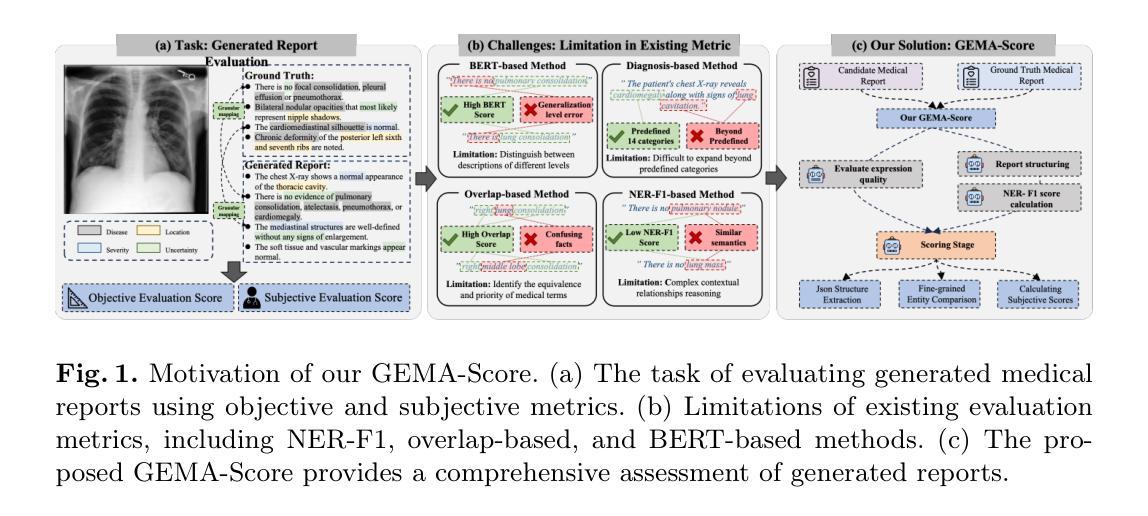
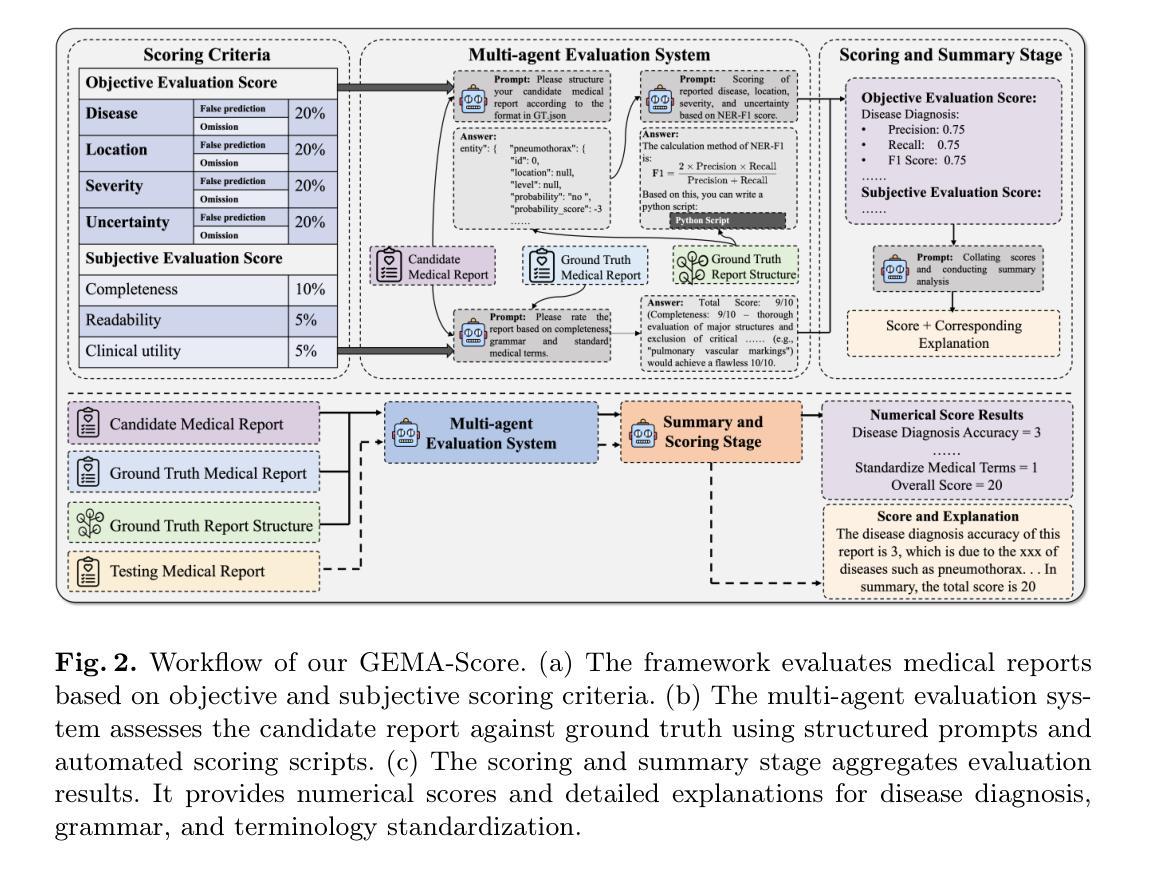
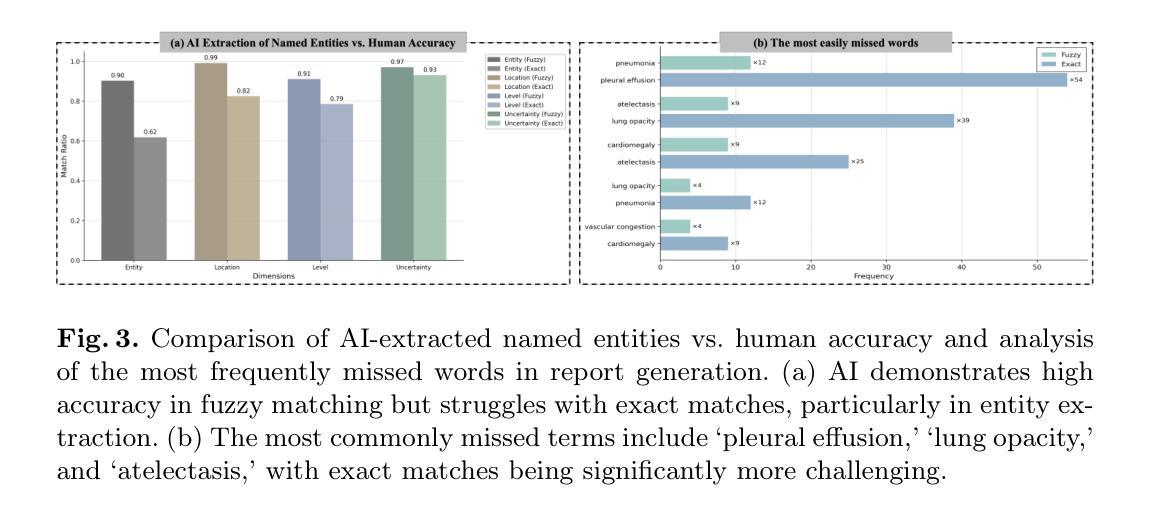
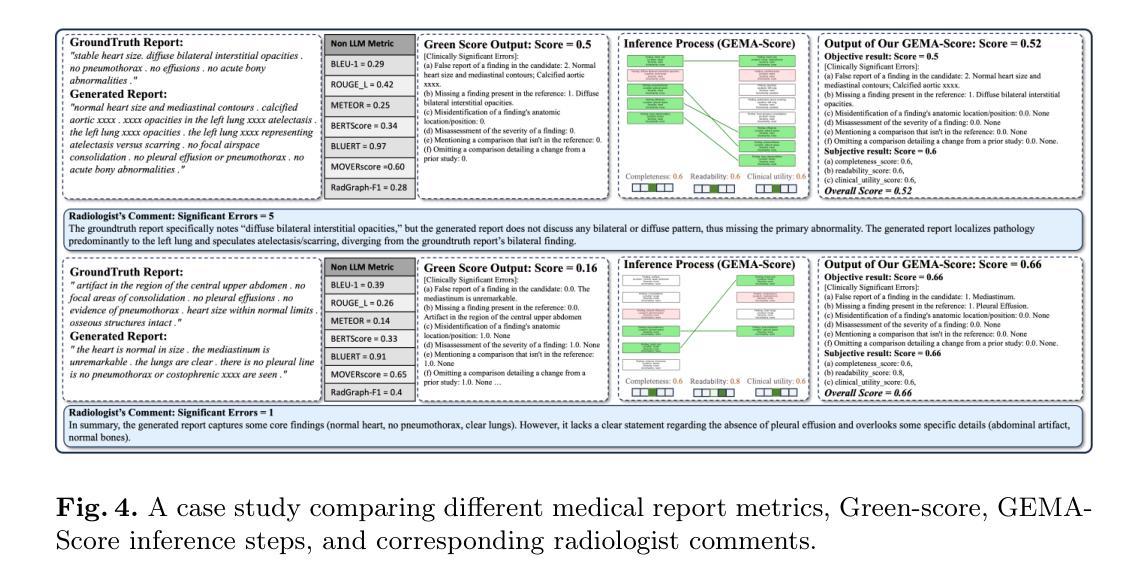
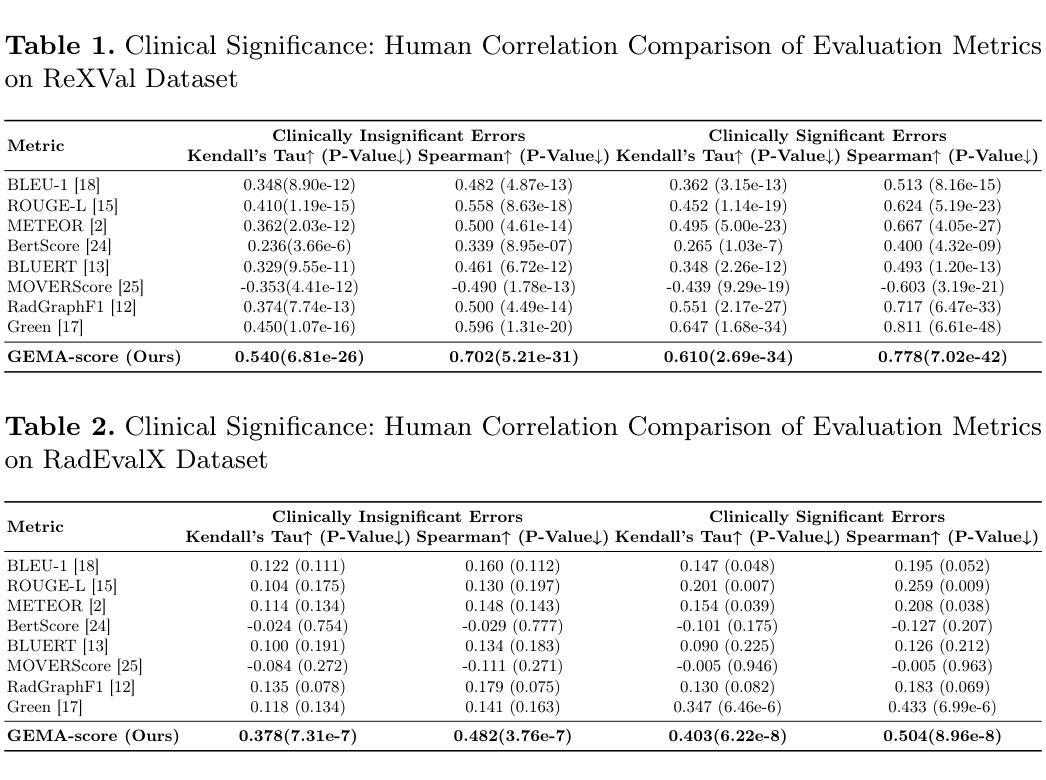
GEMA-Score: Granular Explainable Multi-Agent Score for Radiology Report Evaluation
Authors:Zhenxuan Zhang, Kinhei Lee, Weihang Deng, Huichi Zhou, Zihao Jin, Jiahao Huang, Zhifan Gao, Dominic C Marshall, Yingying Fang, Guang Yang
Automatic medical report generation supports clinical diagnosis, reduces the workload of radiologists, and holds the promise of improving diagnosis consistency. However, existing evaluation metrics primarily assess the accuracy of key medical information coverage in generated reports compared to human-written reports, while overlooking crucial details such as the location and certainty of reported abnormalities. These limitations hinder the comprehensive assessment of the reliability of generated reports and pose risks in their selection for clinical use. Therefore, we propose a Granular Explainable Multi-Agent Score (GEMA-Score) in this paper, which conducts both objective quantification and subjective evaluation through a large language model-based multi-agent workflow. Our GEMA-Score parses structured reports and employs NER-F1 calculations through interactive exchanges of information among agents to assess disease diagnosis, location, severity, and uncertainty. Additionally, an LLM-based scoring agent evaluates completeness, readability, and clinical terminology while providing explanatory feedback. Extensive experiments validate that GEMA-Score achieves the highest correlation with human expert evaluations on a public dataset, demonstrating its effectiveness in clinical scoring (Kendall coefficient = 0.70 for Rexval dataset and Kendall coefficient = 0.54 for RadEvalX dataset). The anonymous project demo is available at: https://github.com/Zhenxuan-Zhang/GEMA_score.
自动生成的医学报告支持临床诊断,减轻了放射科医师的工作量,并有望提高诊断的一致性。然而,现有的评估指标主要评估自动生成报告与人工编写报告相比关键医疗信息覆盖的准确性,却忽视了报告异常的位置和确定性等重要细节。这些局限性阻碍了生成报告的可靠性的全面评估,并给其临床使用选择带来了风险。因此,本文提出了一种细粒度可解释多智能体评分(GEMA-Score),它采用基于大型语言模型的多智能体工作流程进行客观量化和主观评价。我们的GEMA-Score解析结构化报告,通过智能体之间的信息交互交流,利用NER-F1计算来评估疾病诊断、位置、严重程度和不确定性。此外,基于LLM的评分智能体评估完整性、可读性和临床术语,同时提供解释性反馈。大量实验验证,GEMA-Score在公共数据集上与人类专家评价的相关性最高,证明了其在临床评分中的有效性(Rexval数据集的Kendall系数为0.70,RadEvalX数据集的Kendall系数为0.54)。匿名项目演示地址:https://github.com/Zhenxuan-Zhang/GEMA_score。
论文及项目相关链接
Summary
医学报告自动生成技术有助于临床诊断,减轻医生工作量,提高诊断一致性。但现有评估指标主要关注生成的报告是否覆盖关键医学信息,而忽视了异常部位的定位和确定性等重要细节。本文提出一种名为Granular Explainable Multi-Agent Score(GEMA-Score)的评估方法,通过多智能体工作流程进行客观量化和主观评价。该方法能评估疾病诊断、部位、严重程度和不确定性等,同时评价报告的完整性、可读性和临床术语使用。实验证明,GEMA-Score在公共数据集上与人专家评价的关联度最高,可有效用于临床评分。
Key Takeaways
- 医学报告自动生成技术有助于提升临床诊断和治疗效率。
- 当前评估方法主要关注医学信息的覆盖,但忽视了异常部位和确定性等关键细节。
- 提出的GEMA-Score方法结合了客观量化和主观评价。
- GEMA-Score能够评估疾病诊断、部位、严重程度和不确定性。
- 除了评估功能,GEMA-Score还能评价报告的完整性、可读性和临床术语的使用。
- 实验结果显示,GEMA-Score与人的专家评价的关联度最高。
点此查看论文截图

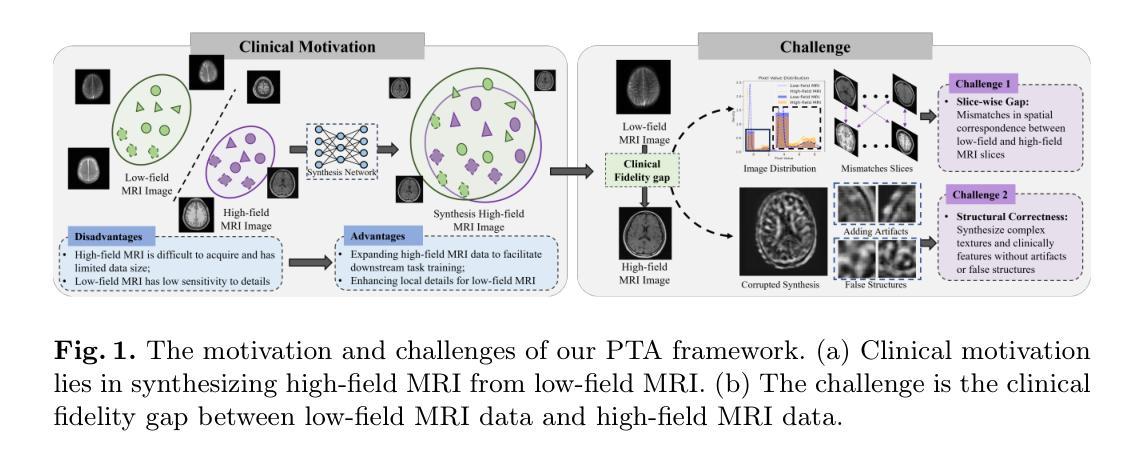
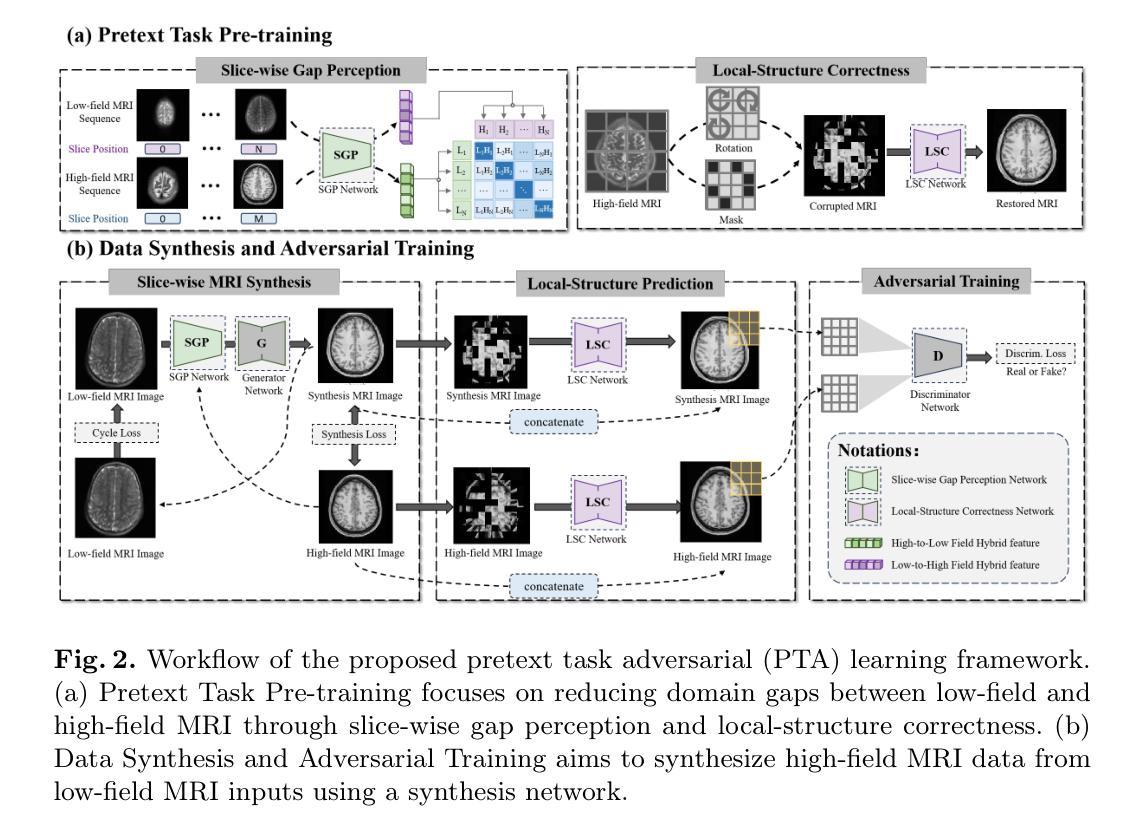
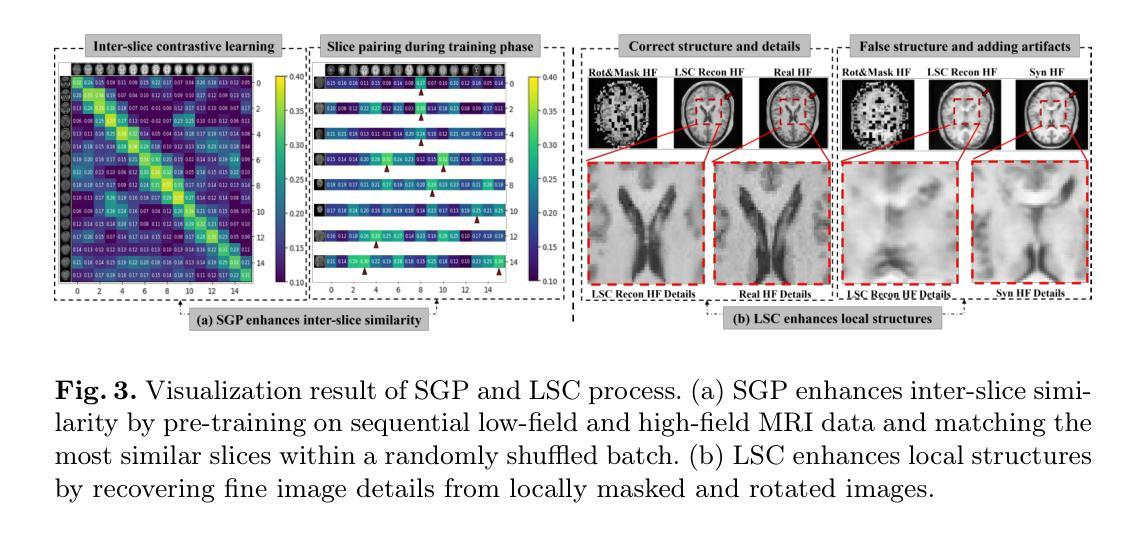
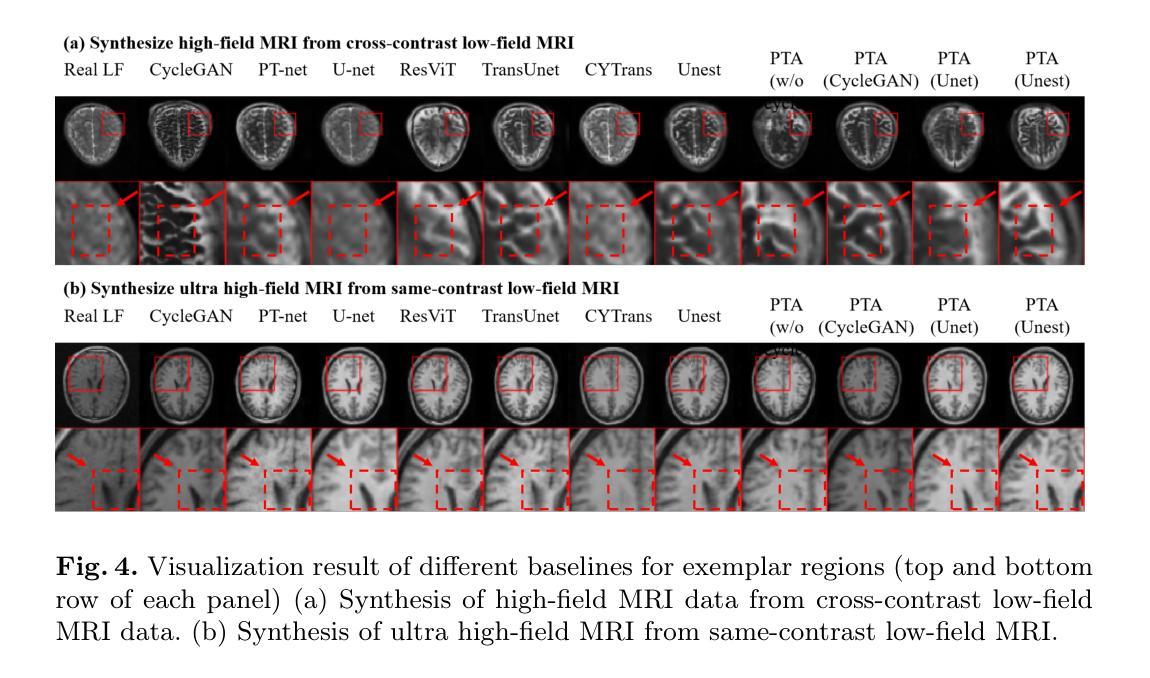
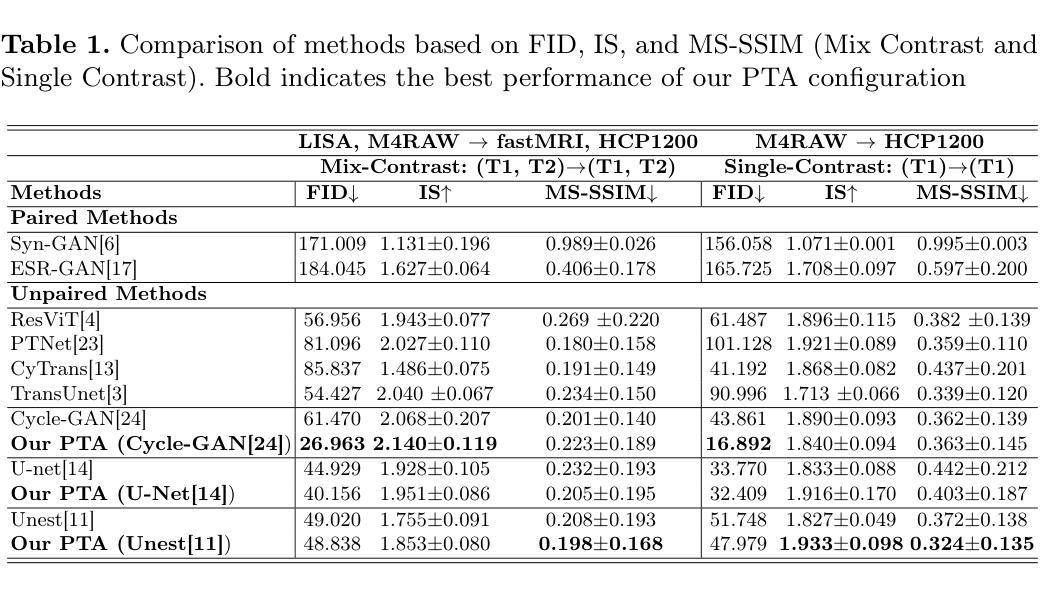
Pretext Task Adversarial Learning for Unpaired Low-field to Ultra High-field MRI Synthesis
Authors:Zhenxuan Zhang, Peiyuan Jing, Coraline Beitone, Jiahao Huang, Zhifan Gao, Guang Yang, Pete Lally
Given the scarcity and cost of high-field MRI, the synthesis of high-field MRI from low-field MRI holds significant potential when there is limited data for training downstream tasks (e.g. segmentation). Low-field MRI often suffers from a reduced signal-to-noise ratio (SNR) and spatial resolution compared to high-field MRI. However, synthesizing high-field MRI data presents challenges. These involve aligning image features across domains while preserving anatomical accuracy and enhancing fine details. To address these challenges, we propose a Pretext Task Adversarial (PTA) learning framework for high-field MRI synthesis from low-field MRI data. The framework comprises three processes: (1) The slice-wise gap perception (SGP) network aligns the slice inconsistencies of low-field and high-field datasets based on contrastive learning. (2) The local structure correction (LSC) network extracts local structures by restoring the locally rotated and masked images. (3) The pretext task-guided adversarial training process introduces additional supervision and incorporates a discriminator to improve image realism. Extensive experiments on low-field to ultra high-field task demonstrate the effectiveness of our method, achieving state-of-the-art performance (16.892 in FID, 1.933 in IS, and 0.324 in MS-SSIM). This enables the generation of high-quality high-field-like MRI data from low-field MRI data to augment training datasets for downstream tasks. The code is available at: https://github.com/Zhenxuan-Zhang/PTA4Unpaired_HF_MRI_SYN.
鉴于高场MRI的稀缺性和成本,当训练下游任务(例如分割)的数据有限时,从低场MRI合成高场MRI具有重要的潜力。与高场MRI相比,低场MRI通常具有较低的信噪比(SNR)和空间分辨率。然而,合成高场MRI数据存在挑战,这涉及到跨域对齐图像特征,同时保留解剖精度并增强细节。为了解决这些挑战,我们提出了一个从低场MRI数据合成高场MRI的Pretext Task Adversarial(PTA)学习框架。该框架包括三个过程:(1)切片级间隙感知(SGP)网络基于对比学习对齐低场和高场数据集的不一致性切片。(2)局部结构校正(LSC)网络通过恢复局部旋转和遮挡的图像来提取局部结构。(3)预训练任务引导对抗训练过程引入了额外的监督并合并了一个鉴别器,以提高图像的真实性。在低场到超高场的广泛实验表明,我们的方法非常有效,达到了最先进的性能(FID为16.892,IS为1.933,MS-SSIM为0.324)。这能够从低场MRI数据生成高质量的高场MRI数据,以扩充下游任务的训练数据集。代码可在以下网址找到:https://github.com/Zhenxuan-Zhang/PTA4Unpaired_HF_MRI_SYN。
论文及项目相关链接
摘要
本文介绍了利用低场MRI数据合成高场MRI数据的方法,以解决高场MRI稀缺和成本高昂的问题。针对低场MRI的信号噪声比和空间分辨率较低的问题,提出了基于对抗学习(PTA)的高场MRI合成框架。该框架包括切片不一致性对齐、局部结构校正和对抗训练三个过程,实现了从低场到高场的MRI数据合成,并达到了领先水平。此技术有助于扩充训练数据集,为下游任务(如分割)提供高质量的高场MRI数据。相关代码已公开。
关键见解
- 利用低场MRI数据合成高场MRI数据,以解决高场MRI资源不足的问题。
- 介绍了PTA学习框架,包括切片不一致性对齐、局部结构校正和对抗训练三个关键过程。
- 通过广泛实验验证了该方法的有效性,达到了领先水平,并提供了具体评价指标(FID=16.892, IS=1.933, MS-SSIM=0.324)。
- 该技术有助于提高训练数据集的质量,为下游任务(如医学图像分割)提供有益支持。
- 公开了相关代码,便于其他研究者使用和改进。
- 此技术对于提高医学图像分析的准确性和效率具有重要意义。
点此查看论文截图

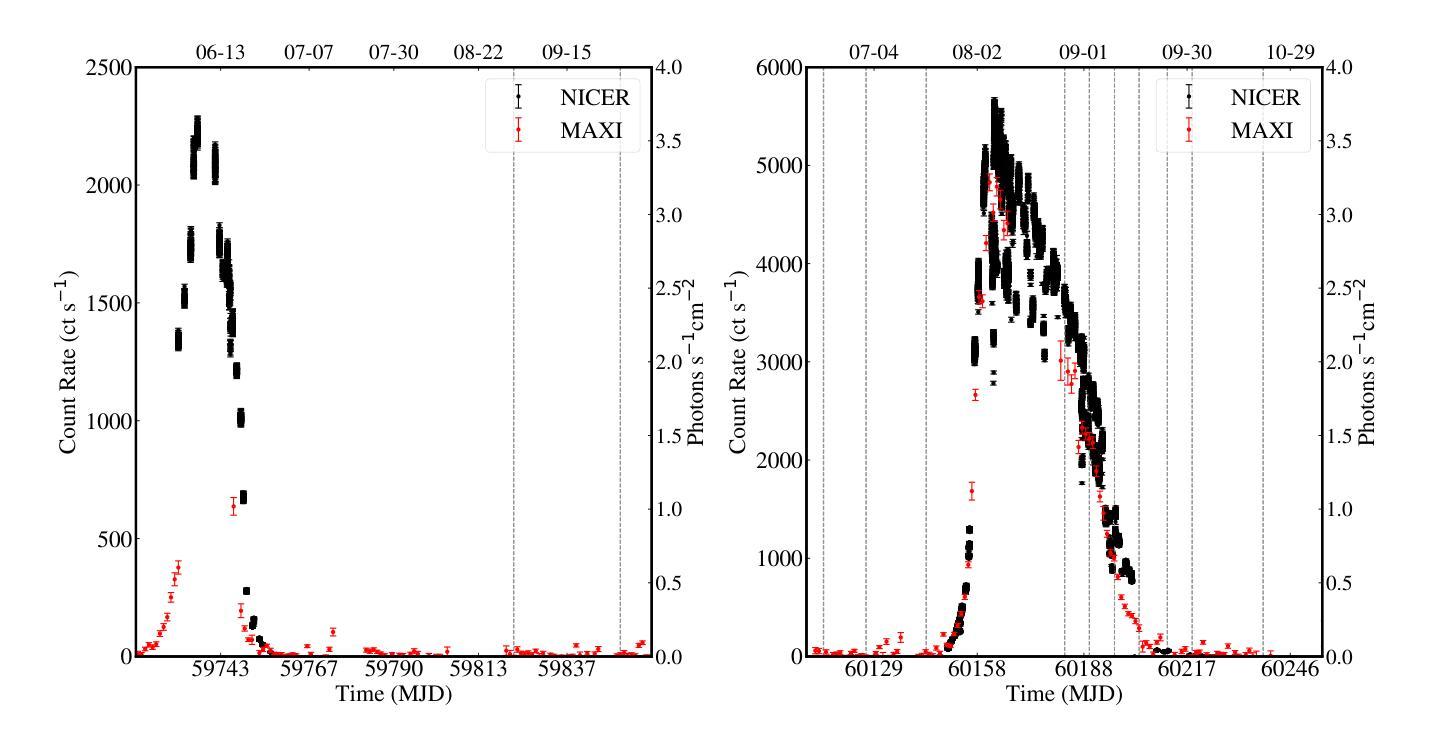
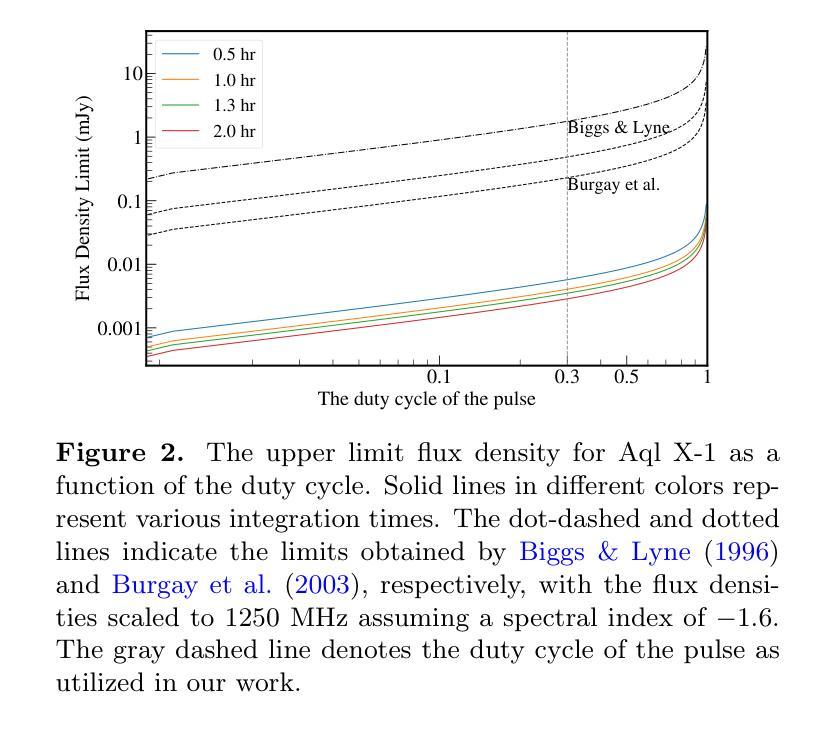
Radio pulse search from Aql X-1
Authors:Long Peng, Zhaosheng Li, Yuanyue Pan, Shanshan Weng, Wengming Yan, Na Wang, Bojun Wang, Shuangqiang Wang
We present 12 observations of the accreting millisecond X-ray pulsar Aql X-1, taken from August 2022 to October 2023 using the Five-hundred-meter Aperture Spherical Radio Telescope at 1250 MHz. These observations covered both the quiescence and X-ray outburst states, as determined by analyzing the X-ray data from the Neutron Star Interior Composition Explorer and the Monitor of All-sky X-ray Image. Periodicity and single-pulse searches were conducted for each observation, but no pulsed signals were detected. The obtained upper limit flux densities are in the range of 2.86-5.73 uJy, which provide the lowest limits to date. We discuss several mechanisms that may prevent detection, suggesting that Aql X-1 may be in the radio-ejection state during quiescence, where the radio pulsed emissions are absorbed by the matter surrounding the system.
我们展示了使用五百年口径球面射电望远镜在1250 MHz频率下从2022年8月到2023年10月对增亮型毫秒X射线脉冲星Aql X-1进行的12次观测结果。这些观测涵盖了静息态和X射线爆发态,这是通过分析来自中子星内部结构探测器和全天空X射线图像监测器的X射线数据来确定的。我们对每次观测进行了周期性和单脉冲搜索,但没有检测到脉冲信号。所获得的流量密度上限范围为2.86-5.73微吉,这是迄今为止的最低值。我们讨论了几种可能导致无法检测到的机制,并提出Aql X-1可能在静息态时处于射电喷射状态,此时射电脉冲辐射被系统周围的物质吸收。
论文及项目相关链接
Summary
使用射电望远镜观测了射电毫秒脉冲星Aql X-1的十二次观测数据,覆盖其平静期与X射线爆发状态。周期性及单一脉冲搜索均未发现脉冲信号,获得的上限流量密度范围在2.86-5.73 uJy之间,为目前最低值。讨论了可能阻碍探测的机制,提出Aql X-1在平静期可能处于射电喷射状态,射电脉冲排放被系统周围物质吸收。
Key Takeaways
- 使用射电望远镜观测了Aql X-1的十二次数据。
- 观测覆盖了Aql X-1的平静期和X射线爆发状态。
- 搜索过程中未发现周期性或单一脉冲信号。
- 获得的上限流量密度是迄今为止的最低值,范围在2.86-5.73 uJy之间。
- 讨论了可能阻碍射电信号探测的机制。
- 提出Aql X-1在平静期可能处于射电喷射状态。
点此查看论文截图


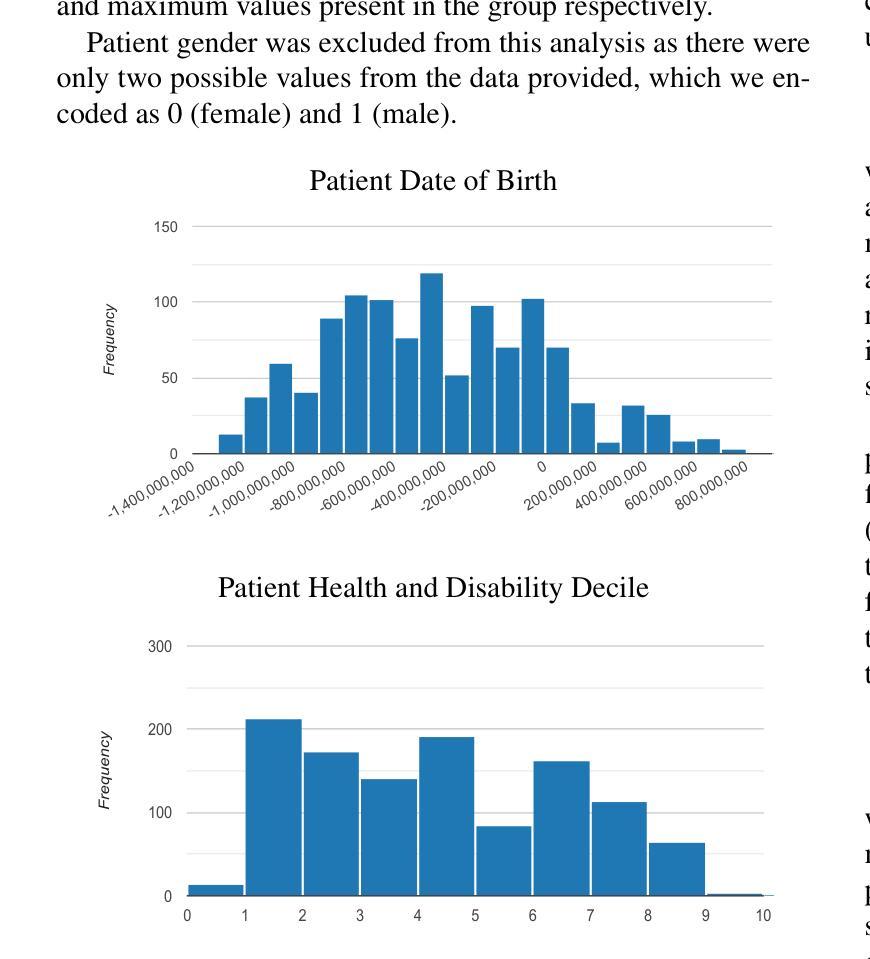
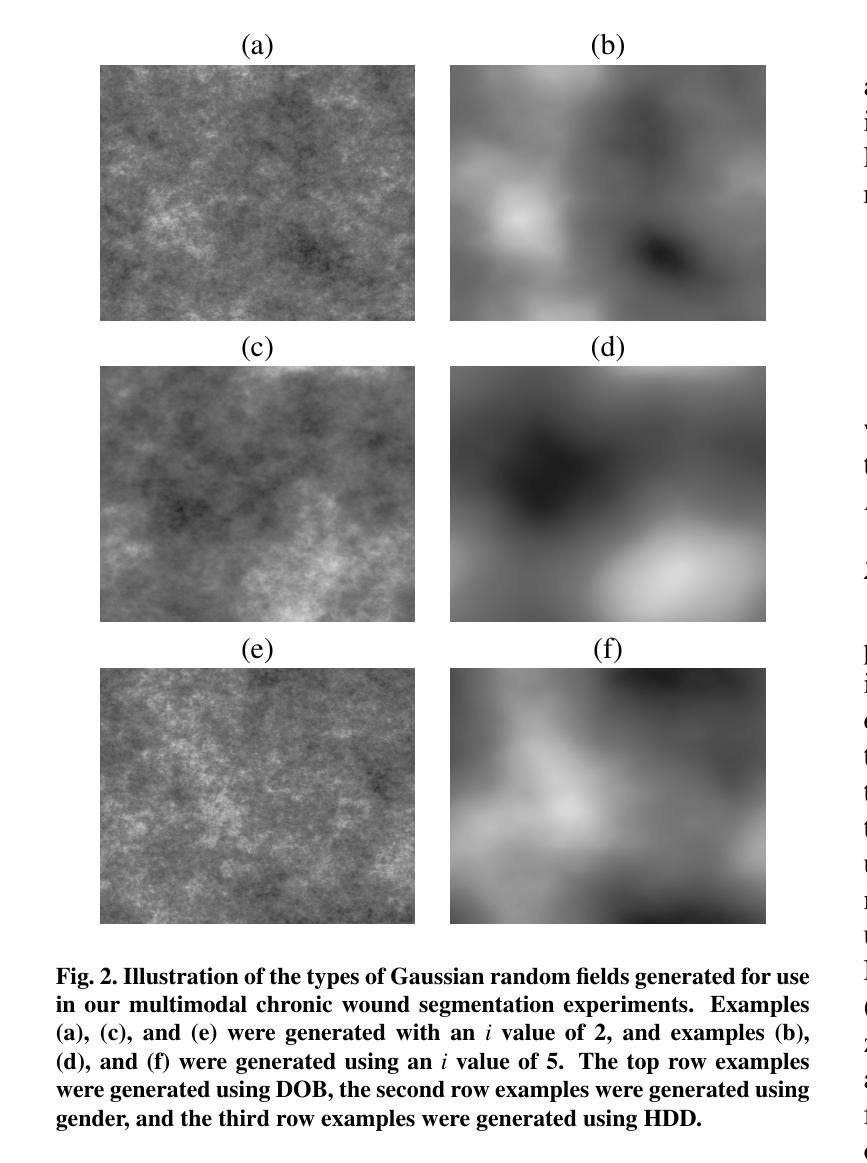
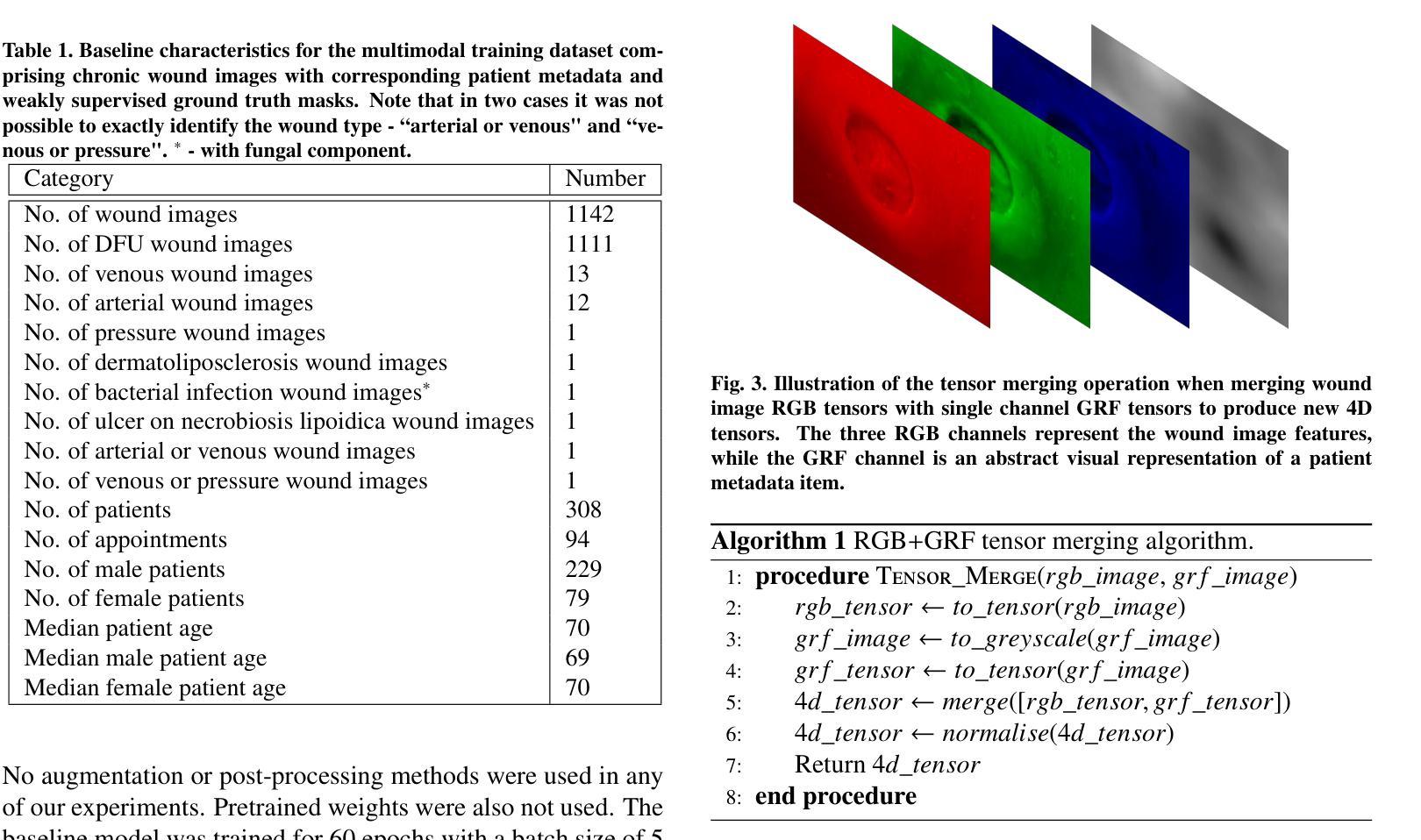
Gaussian Random Fields as an Abstract Representation of Patient Metadata for Multimodal Medical Image Segmentation
Authors:Bill Cassidy, Christian McBride, Connah Kendrick, Neil D. Reeves, Joseph M. Pappachan, Shaghayegh Raad, Moi Hoon Yap
The growing rate of chronic wound occurrence, especially in patients with diabetes, has become a concerning trend in recent years. Chronic wounds are difficult and costly to treat, and have become a serious burden on health care systems worldwide. Chronic wounds can have devastating consequences for the patient, with infection often leading to reduced quality of life and increased mortality risk. Innovative deep learning methods for the detection and monitoring of such wounds have the potential to reduce the impact to both patient and clinician. We present a novel multimodal segmentation method which allows for the introduction of patient metadata into the training workflow whereby the patient data are expressed as Gaussian random fields. Our results indicate that the proposed method improved performance when utilising multiple models, each trained on different metadata categories. Using the Diabetic Foot Ulcer Challenge 2022 test set, when compared to the baseline results (intersection over union = 0.4670, Dice similarity coefficient = 0.5908) we demonstrate improvements of +0.0220 and +0.0229 for intersection over union and Dice similarity coefficient respectively. This paper presents the first study to focus on integrating patient data into a chronic wound segmentation workflow. Our results show significant performance gains when training individual models using specific metadata categories, followed by average merging of prediction masks using distance transforms. All source code for this study is available at: https://github.com/mmu-dermatology-research/multimodal-grf
近年来,慢性伤口的发生率,特别是在糖尿病患者中,呈现出令人担忧的增长趋势。慢性伤口的治疗既困难又昂贵,已成为全球医疗系统面临的一项严重负担。对于患者而言,慢性伤口可能产生灾难性的后果,感染常常导致生活质量下降和死亡风险增加。针对此类伤口的检测和监测,创新的深度学习方法具有减轻患者和临床医生负担的潜力。我们提出了一种新型的多模态分割方法,允许将患者元数据引入训练流程中,患者数据以高斯随机场的形式表达。我们的结果表明,在利用多个模型时,所提出的方法提高了性能,每个模型都在不同的元数据集上进行训练。在Diabetic Foot Ulcer Challenge 2022测试集上,与基线结果相比(交并比=0.4670,狄氏相似系数=0.5908),我们的交并比和狄氏相似系数分别提高了+0.0220和+0.0229。本文是首次专注于将患者数据集成到慢性伤口分割流程中的研究。我们的结果表明,当使用特定的元数据集对单个模型进行训练,然后采用距离变换对预测掩膜进行平均合并时,可以获得显著的性能提升。该研究的所有源代码可在以下网址找到:https://github.com/mmu-dermatology-research/multimodal-grf 。
论文及项目相关链接
Summary
本文关注慢性伤口,特别是糖尿病患者慢性伤口的日益增长趋势,提出一种新颖的多模态分割方法,该方法将患者元数据引入训练流程,并以高斯随机场的形式表达。在糖尿病足溃疡挑战2022测试集上的结果显示,该方法相较于基线结果有所提高。本文集成了患者数据到慢性伤口分割流程中,并通过特定元数据集训练模型再平均合并预测掩膜的方式,取得了显著的性能提升。
Key Takeaways
- 慢性伤口,尤其是糖尿病患者的慢性伤口,其发生率呈增长趋势,成为全球医疗系统的严重负担。
- 慢性伤口的治疗困难且费用高昂,对患者的生活质量和生命健康产生严重影响。
- 深度学习在检测与监控慢性伤口中具有巨大潜力。
- 提出一种新颖的多模态分割方法,将患者元数据引入训练流程。
- 该方法通过高斯随机场表达患者数据,并在多个模型上取得良好性能。
- 在糖尿病足溃疡挑战2022测试集上,该方法相较于基线结果有所提升,显示出方法的有效性。
点此查看论文截图

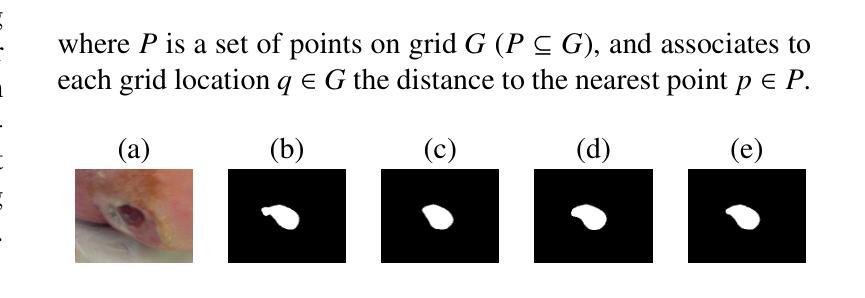
Partially Supervised Unpaired Multi-Modal Learning for Label-Efficient Medical Image Segmentation
Authors:Lei Zhu, Yanyu Xu, Huazhu Fu, Xinxing Xu, Rick Siow Mong Goh, Yong Liu
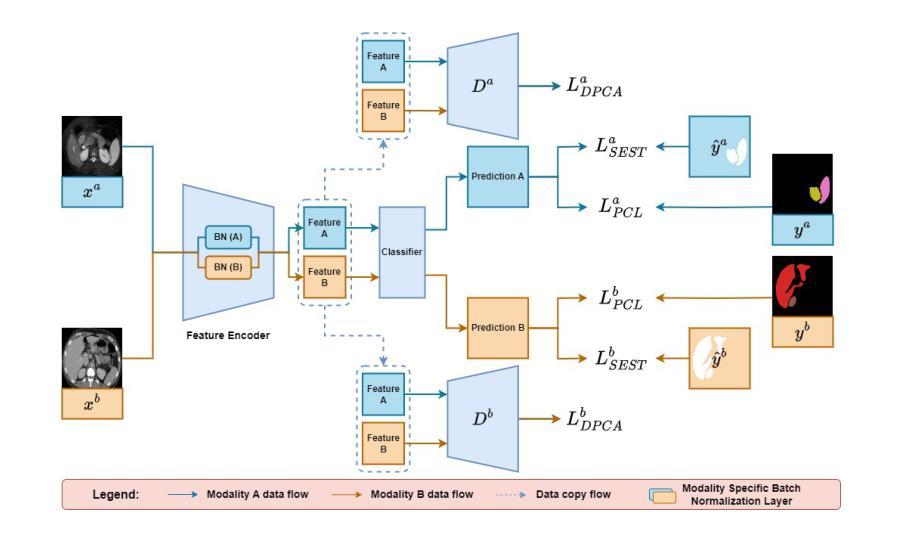
Unpaired Multi-Modal Learning (UMML) which leverages unpaired multi-modal data to boost model performance on each individual modality has attracted a lot of research interests in medical image analysis. However, existing UMML methods require multi-modal datasets to be fully labeled, which incurs tremendous annotation cost. In this paper, we investigate the use of partially labeled data for label-efficient unpaired multi-modal learning, which can reduce the annotation cost by up to one half. We term the new learning paradigm as Partially Supervised Unpaired Multi-Modal Learning (PSUMML) and propose a novel Decomposed partial class adaptation with snapshot Ensembled Self-Training (DEST) framework for it. Specifically, our framework consists of a compact segmentation network with modality specific normalization layers for learning with partially labeled unpaired multi-modal data. The key challenge in PSUMML lies in the complex partial class distribution discrepancy due to partial class annotation, which hinders effective knowledge transfer across modalities. We theoretically analyze this phenomenon with a decomposition theorem and propose a decomposed partial class adaptation technique to precisely align the partially labeled classes across modalities to reduce the distribution discrepancy. We further propose a snapshot ensembled self-training technique to leverage the valuable snapshot models during training to assign pseudo-labels to partially labeled pixels for self-training to boost model performance. We perform extensive experiments under different scenarios of PSUMML for two medical image segmentation tasks, namely cardiac substructure segmentation and abdominal multi-organ segmentation. Our framework outperforms existing methods significantly.
无配对多模态学习(UMML)利用无配对的多模态数据来提升每个独立模态的模型性能,在医学图像分析领域引起了大量的研究兴趣。然而,现有的UMML方法要求多模态数据集完全标注,这产生了巨大的标注成本。在本文中,我们研究了利用部分标注数据进行标签高效的无配对多模态学习,这可以将标注成本降低一半。我们将这种新的学习范式称为部分监督无配对多模态学习(PSUMML),并提出了一个名为分解部分类适应与快照集成自训练(DEST)的新框架。具体来说,我们的框架由一个紧凑的分割网络组成,该网络具有针对部分标注无配对多模态数据学习的模态特定归一化层。PSUMML的关键挑战在于部分类分布差异复杂,这是由于部分类标注造成的,阻碍了跨模态的有效知识转移。我们通过分解定理对其进行理论分析,并提出了一种分解的部分类适应技术,以精确对齐部分标注的跨模态类别,减少分布差异。我们进一步提出了一种快照集成自训练技术,以利用训练过程中的有价值的快照模型,对部分标注的像素进行自训练,以提高模型性能。我们在两个不同的PSUMML场景下进行了大量实验,涉及两个医学图像分割任务,即心脏子结构分割和腹部多器官分割。我们的框架显著优于现有方法。
论文及项目相关链接
PDF Accepted to MLMI 2024
摘要
该研究探索了利用部分标记数据实现标签效率低的非配对多模态学习(PSUMML),可节省一半的标注成本。研究提出了名为分解部分类别自适应快照集成自训练(DEST)的新型框架。框架具有模态特定归一化层的紧凑分割网络,用于处理部分标记的非配对多模态数据。PSUMML的主要挑战在于由于部分类别标注造成的复杂部分类别分布差异,阻碍了跨模态的有效知识转移。研究通过分解定理进行理论分析,并提出了精确对齐跨模态部分标记类别的分解部分类别自适应技术,以减少分布差异。此外,还提出了快照集成自训练技术,利用训练过程中的快照模型对部分标记像素分配伪标签进行自训练,以提高模型性能。在不同PSUMML场景下的两个医学图像分割任务(心脏子结构分割和腹部多器官分割)的实验表明,该框架显著优于现有方法。
要点摘要
- 研究引入了部分监督的非配对多模态学习(PSUMML),允许使用部分标记的数据来减少标注成本。
- 提出了一种名为DEST的新型框架,利用紧凑的分割网络和模态特定归一化层处理PSUMML中的数据。
- PSUMML面临的主要挑战是部分类别标注引起的复杂分布差异。
- 通过分解定理分析这一问题,并提出分解部分类别自适应技术来解决跨模态的部分标记类别对齐问题。
- 采用快照集成自训练技术,利用训练过程中的快照模型进行自训练,提高模型性能。
- 在医学图像分割任务上进行了大量实验验证DEST框架的有效性。
点此查看论文截图

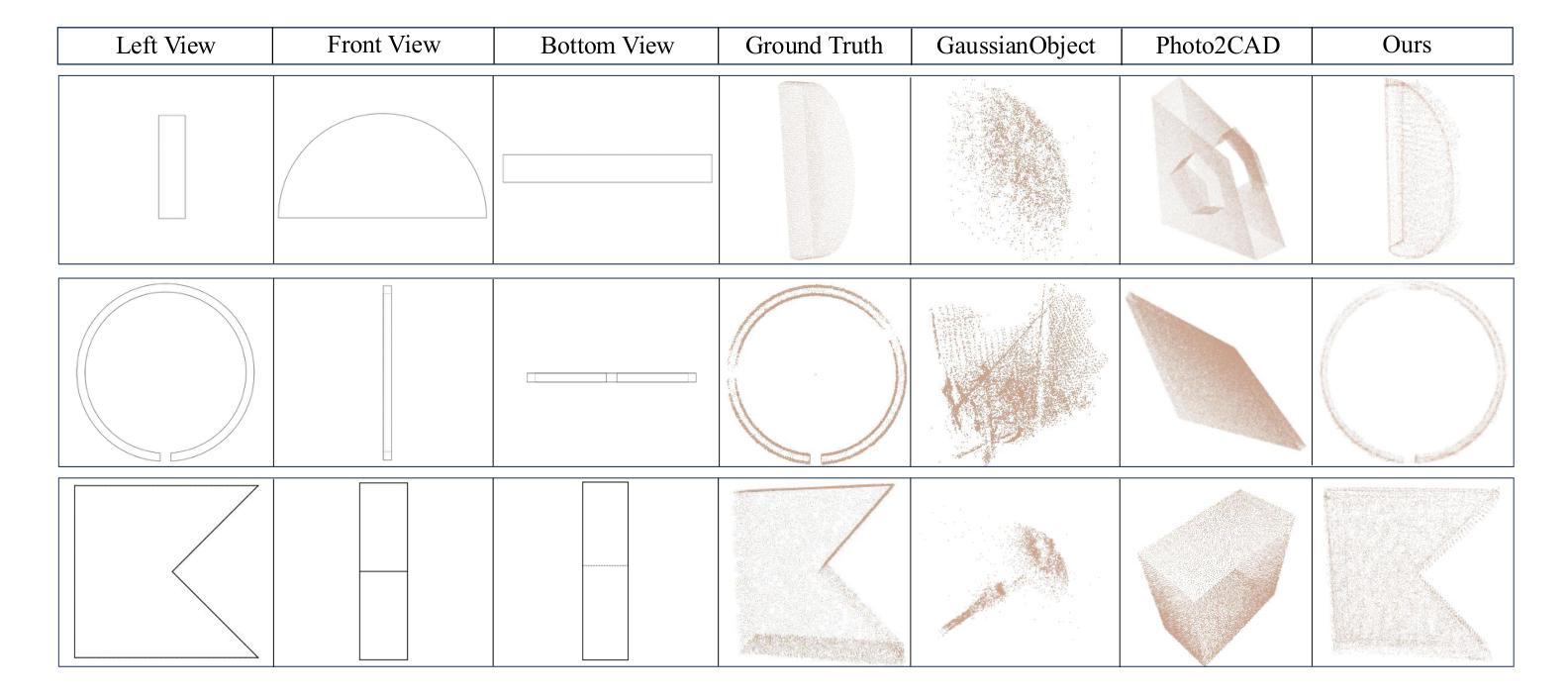
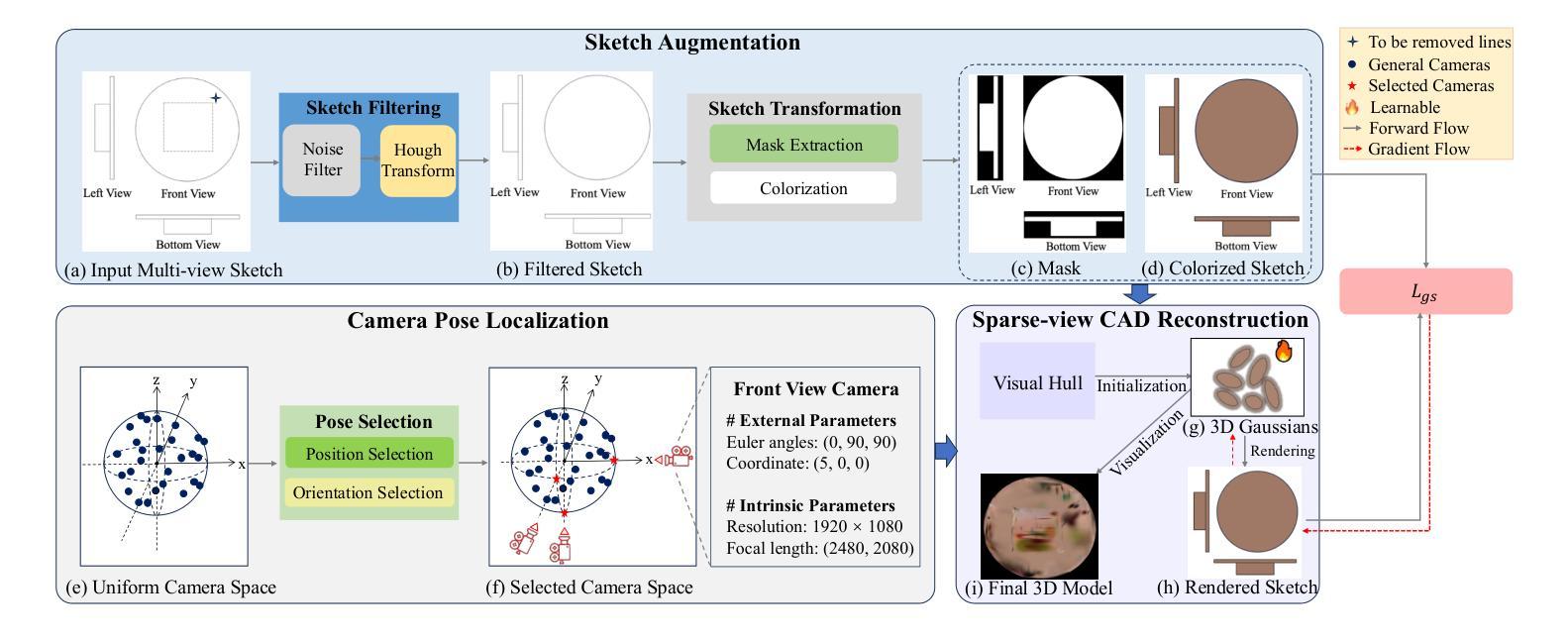
GaussianCAD: Robust Self-Supervised CAD Reconstruction from Three Orthographic Views Using 3D Gaussian Splatting
Authors:Zheng Zhou, Zhe Li, Bo Yu, Lina Hu, Liang Dong, Zijian Yang, Xiaoli Liu, Ning Xu, Ziwei Wang, Yonghao Dang, Jianqin Yin
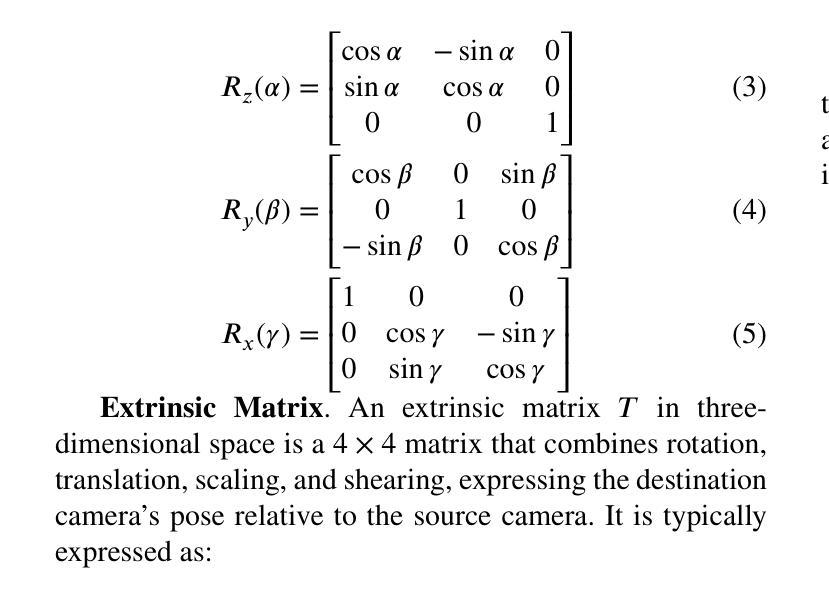
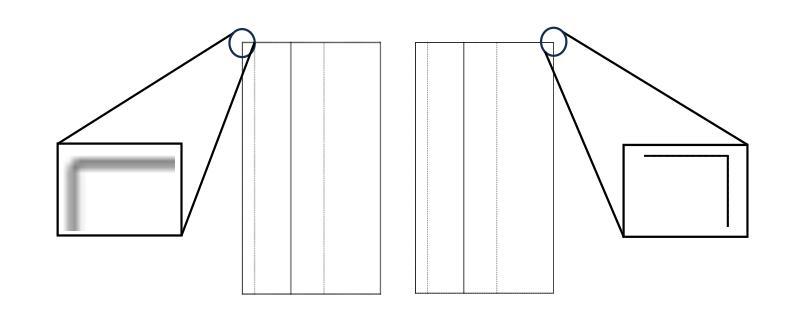
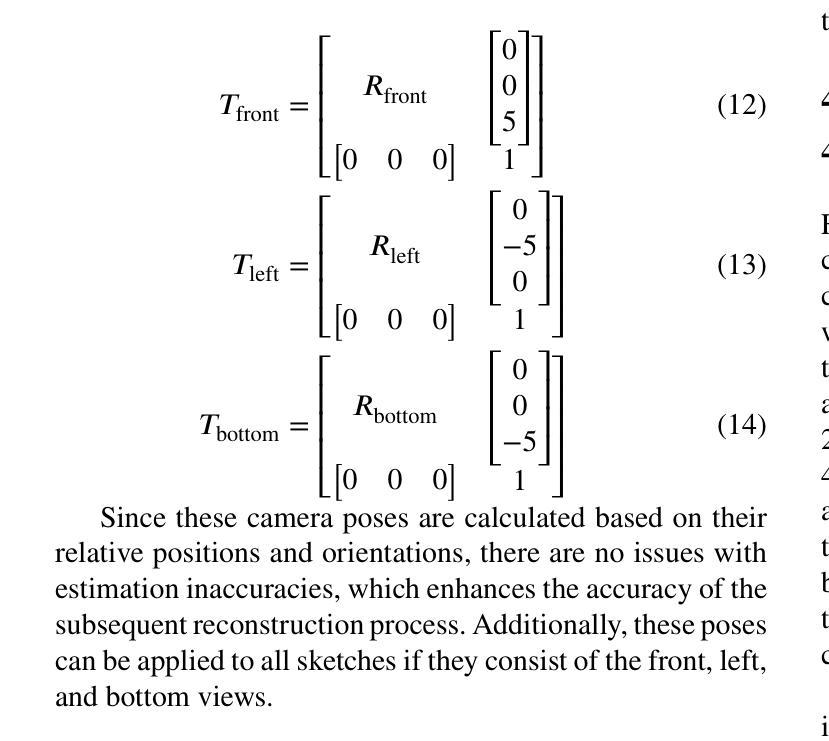
The automatic reconstruction of 3D computer-aided design (CAD) models from CAD sketches has recently gained significant attention in the computer vision community. Most existing methods, however, rely on vector CAD sketches and 3D ground truth for supervision, which are often difficult to be obtained in industrial applications and are sensitive to noise inputs. We propose viewing CAD reconstruction as a specific instance of sparse-view 3D reconstruction to overcome these limitations. While this reformulation offers a promising perspective, existing 3D reconstruction methods typically require natural images and corresponding camera poses as inputs, which introduces two major significant challenges: (1) modality discrepancy between CAD sketches and natural images, and (2) difficulty of accurate camera pose estimation for CAD sketches. To solve these issues, we first transform the CAD sketches into representations resembling natural images and extract corresponding masks. Next, we manually calculate the camera poses for the orthographic views to ensure accurate alignment within the 3D coordinate system. Finally, we employ a customized sparse-view 3D reconstruction method to achieve high-quality reconstructions from aligned orthographic views. By leveraging raster CAD sketches for self-supervision, our approach eliminates the reliance on vector CAD sketches and 3D ground truth. Experiments on the Sub-Fusion360 dataset demonstrate that our proposed method significantly outperforms previous approaches in CAD reconstruction performance and exhibits strong robustness to noisy inputs.
从计算机辅助设计(CAD)草图自动重建3D计算机辅助设计(CAD)模型最近在计算机视觉领域引起了广泛关注。然而,大多数现有方法都依赖于矢量CAD草图和3D真实值进行监管,这在工业应用中往往难以获得,并且对噪声输入很敏感。为了解决这些局限性,我们提出将CAD重建视为稀疏视图3D重建的一个特定实例。虽然这种重新表述提供了一个有前景的视角,但现有的3D重建方法通常需要自然图像和相应的相机姿态作为输入,这带来了两大挑战:(1)CAD草图与自然图像之间的模态差异,以及(2)为CAD草图准确估计相机姿态的困难。为了解决这些问题,我们首先将CAD草图转换为类似于自然图像的表示形式,并提取相应的掩模。接下来,我们手动计算正射视图的相机姿态,以确保在3D坐标系内的准确对齐。最后,我们采用定制的稀疏视图3D重建方法,从对齐的正射视图中实现高质量重建。通过利用栅格CAD草图进行自监督,我们的方法消除了对矢量CAD草图和3D真实值的依赖。在Sub-Fusion360数据集上的实验表明,我们提出的方法在CAD重建性能上显著优于以前的方法,并对嘈杂输入表现出强大的稳健性。
论文及项目相关链接
Summary
该研究关注于将计算机视觉应用于计算机辅助设计(CAD)模型的自动重建问题,解决了从CAD草图重构三维模型所面临的困难。该方法借鉴了稀疏视图的三维重建思想,利用矢量CAD草图以及相机姿态数据解决重构难题,有效消除了噪声输入干扰,并将模型泛化应用于新的草图输入。最终通过Sub-Fusion360数据集的实验验证,该方法的性能优于现有方法,具有更强的鲁棒性。
Key Takeaways
- 研究针对CAD草图自动重建三维模型的问题进行了深入探讨。
- 研究提出了一种基于稀疏视图的三维重建方法来重建CAD模型,避免了现有方法依赖于矢量CAD草图以及真实三维模型的困难。
- 研究解决了CAD草图与真实图像之间的模态差异问题以及相机姿态估计的难题。通过将CAD草图转化为与自然图像相似的表现形式来解决这些问题。
- 方法的核心步骤包括:草图到自然图像形式的转换、提取对应掩膜、手动计算相机姿态以确保在三维坐标系中的准确对齐以及使用定制的稀疏视图三维重建方法实现高质量重建。
- 方法借助自监督技术使用光栅化CAD草图来训练模型,大大提升了模型对于噪声的鲁棒性。
点此查看论文截图

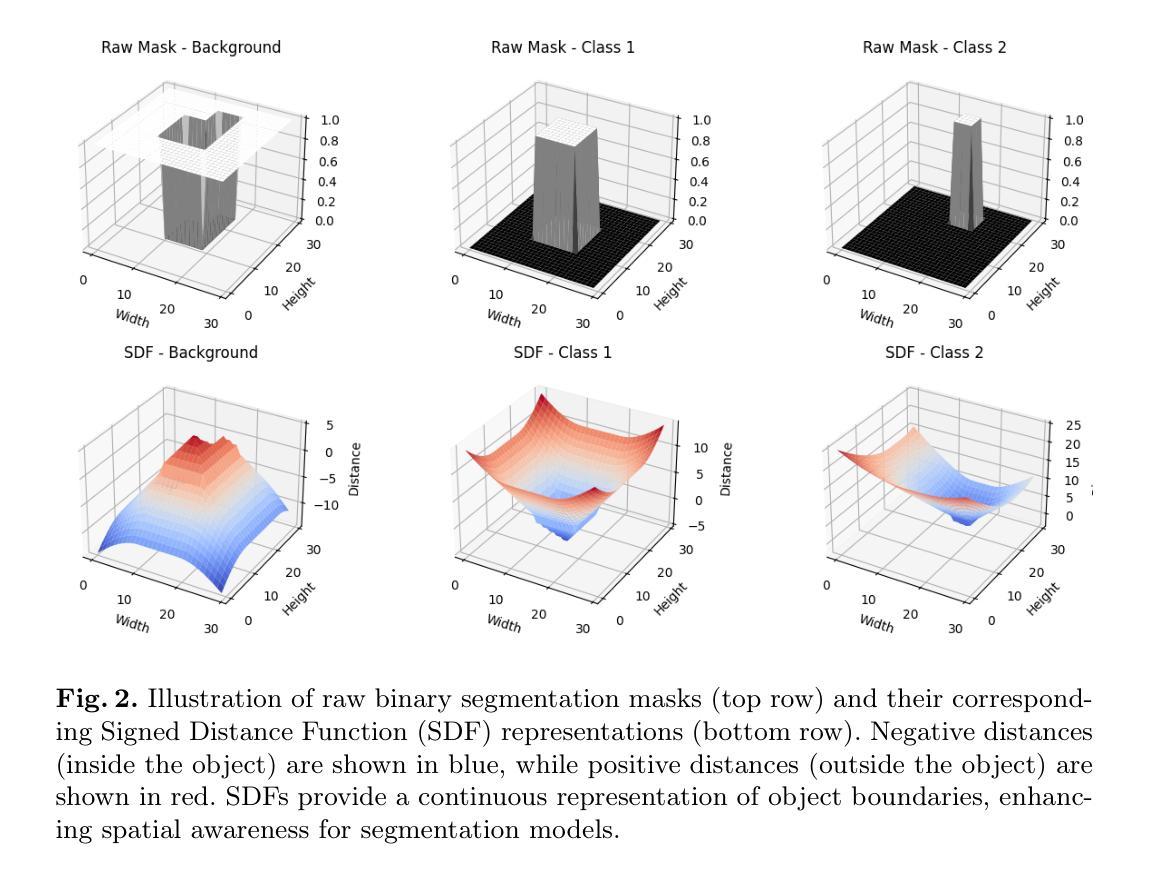
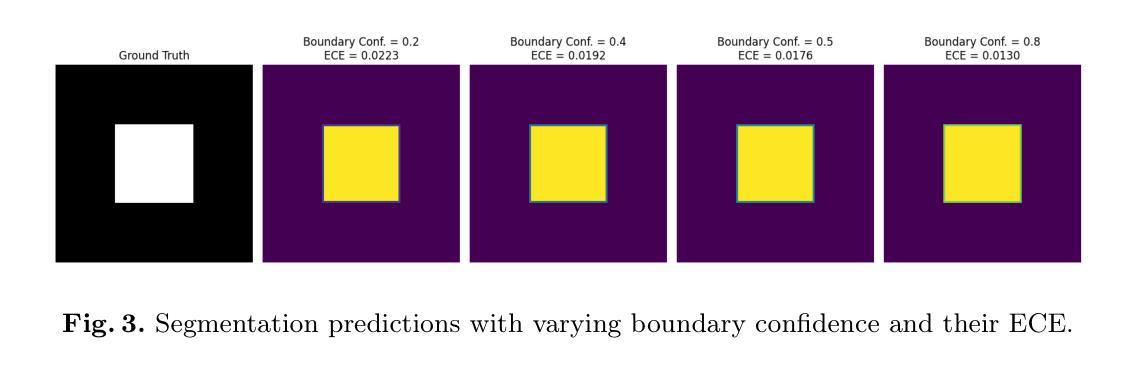
We Care Each Pixel: Calibrating on Medical Segmentation Model
Authors:Wenhao Liang, Wei Zhang, Yue Lin, Miao Xu, Olaf Maennel, Weitong Chen
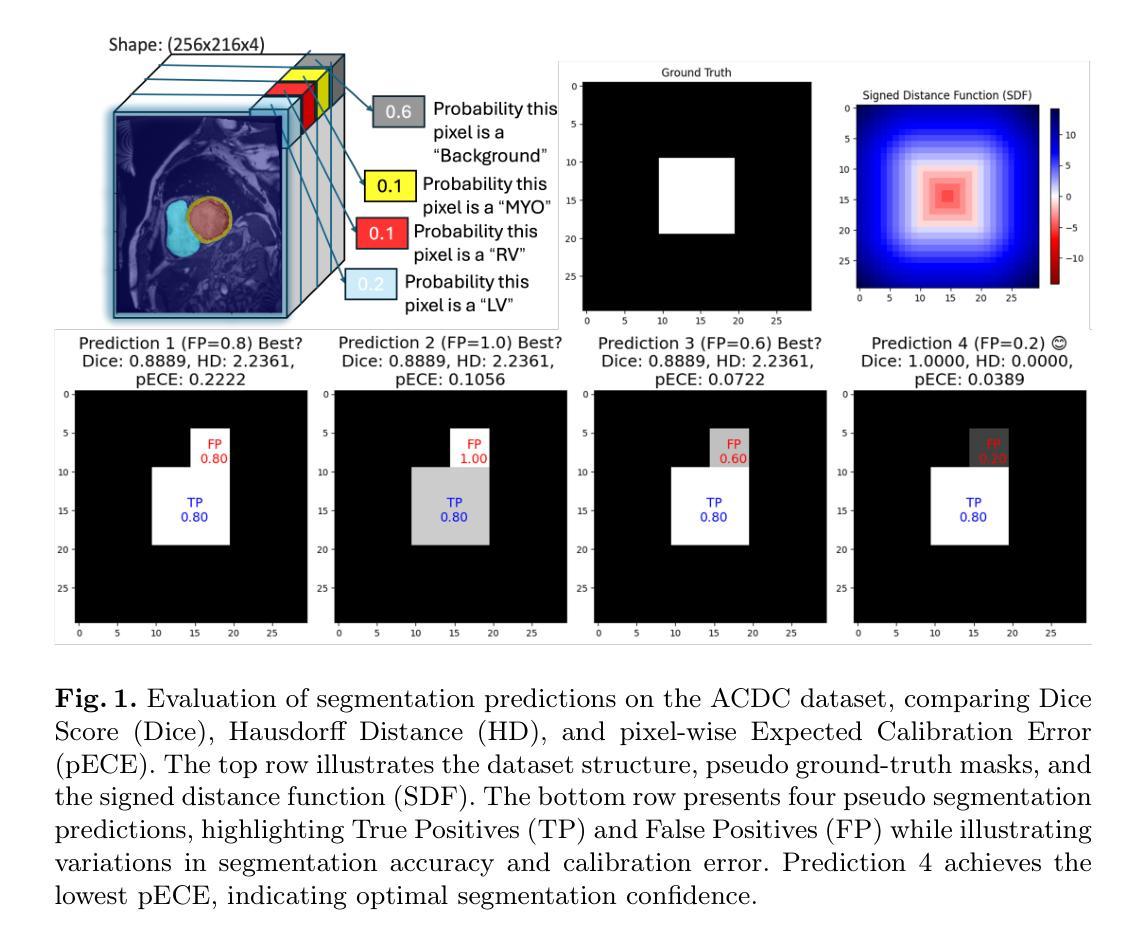
Medical image segmentation is fundamental for computer-aided diagnostics, providing accurate delineation of anatomical structures and pathological regions. While common metrics such as Accuracy, DSC, IoU, and HD primarily quantify spatial agreement between predictions and ground-truth labels, they do not assess the calibration quality of segmentation models, which is crucial for clinical reliability. To address this limitation, we propose pixel-wise Expected Calibration Error (pECE), a novel metric that explicitly measures miscalibration at the pixel level, thereby ensuring both spatial precision and confidence reliability. We further introduce a morphological adaptation strategy that applies morphological operations to ground-truth masks before computing calibration losses, particularly benefiting margin-based losses such as Margin SVLS and NACL. Additionally, we present the Signed Distance Calibration Loss (SDC), which aligns boundary geometry with calibration objectives by penalizing discrepancies between predicted and ground-truth signed distance functions (SDFs). Extensive experiments demonstrate that our method not only enhances segmentation performance but also improves calibration quality, yielding more trustworthy confidence estimates. Code is available at: https://github.com/EagleAdelaide/SDC-Loss.
医学图像分割对于计算机辅助诊断至关重要,它提供了准确的解剖结构和病理区域的描绘。虽然准确度、DSC、IoU和HD等常见指标主要量化预测和真实标签之间的空间一致性,但它们并没有评估分割模型的校准质量,这对于临床可靠性至关重要。为了解决这一局限性,我们提出了像素级期望校准误差(pECE)这一新指标,该指标明确测量像素级别的校准误差,从而确保空间精度和置信度可靠性。我们还引入了一种形态学适应策略,即对真实标签掩膜进行形态学操作,然后再计算校准损失,这对基于边距的损失(如Margin SVLS和NACL)特别有益。此外,我们提出了符号距离校准损失(SDC),它通过惩罚预测和真实标签符号距离函数(SDFs)之间的差异,将边界几何与校准目标对齐。大量实验表明,我们的方法不仅提高了分割性能,还提高了校准质量,产生了更可靠的置信度估计。代码可在:[https://github.com/EagleAdelaide/SDC-Loss获取。](https://github.com/EagleAdelaide/SDC-Loss%E8%8E%B7%E5 %8F%96%E3%80%82)
论文及项目相关链接
PDF Under Reviewing
Summary
医学图像分割对于计算机辅助诊断至关重要,它为解剖结构和病理区域的准确界定提供了依据。针对现有评估指标无法全面评估模型校准质量的问题,本文提出了像素级期望校准误差(pECE)这一新指标,该指标能够明确测量像素级别的误校准情况,从而确保空间精度和置信度可靠性。此外,本文还介绍了形态学适应策略,通过形态学操作对真实标签掩膜进行处理以计算校准损失,对基于边距的损失如Margin SVLS和NACL特别有益。同时,本文提出了符号距离校准损失(SDC),它通过惩罚预测与真实标签的符号距离函数之间的差异来对齐边界几何与校准目标。实验证明,该方法不仅提高了分割性能,还提高了校准质量,产生了更可靠的置信度估计。
Key Takeaways
- 医学图像分割对计算机辅助诊断至关重要,用于准确界定解剖结构和病理区域。
- 现有评估指标如Accuracy、DSC、IoU和HD主要关注预测与真实标签的空间一致性,但无法评估模型的校准质量,这是临床可靠性的关键。
- 像素级期望校准误差(pECE)是一种新指标,用于明确测量像素级别的误校准情况,确保空间精度和置信度可靠性。
- 形态学适应策略通过形态学操作处理真实标签掩膜来计算校准损失,特别适用于基于边距的损失。
- 符号距离校准损失(SDC)通过惩罚预测与真实标签的符号距离函数差异来增强边界几何与校准目标的对齐。
- 本文方法不仅提高了医学图像分割的性能,还提高了模型的校准质量。
点此查看论文截图

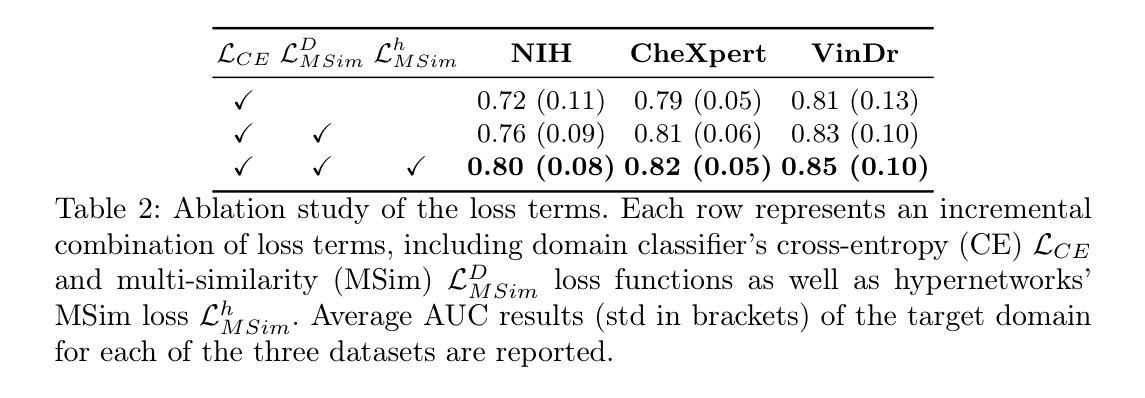
HyDA: Hypernetworks for Test Time Domain Adaptation in Medical Imaging Analysis
Authors:Doron Serebro, Tammy Riklin-Raviv
Medical imaging datasets often vary due to differences in acquisition protocols, patient demographics, and imaging devices. These variations in data distribution, known as domain shift, present a significant challenge in adapting imaging analysis models for practical healthcare applications. Most current domain adaptation (DA) approaches aim either to align the distributions between the source and target domains or to learn an invariant feature space that generalizes well across all domains. However, both strategies require access to a sufficient number of examples, though not necessarily annotated, from the test domain during training. This limitation hinders the widespread deployment of models in clinical settings, where target domain data may only be accessible in real time. In this work, we introduce HyDA, a novel hypernetwork framework that leverages domain characteristics rather than suppressing them, enabling dynamic adaptation at inference time. Specifically, HyDA learns implicit domain representations and uses them to adjust model parameters on-the-fly, effectively interpolating to unseen domains. We validate HyDA on two clinically relevant applications - MRI brain age prediction and chest X-ray pathology classification - demonstrating its ability to generalize across tasks and modalities. Our code is available at TBD.
医学成像数据集往往由于采集协议、患者特征和成像设备等方面的差异而有所不同。数据分布的这些变化被称为域偏移,在为实际医疗保健应用适应成像分析模型时,这构成了重大挑战。当前大多数领域适应(DA)方法旨在对齐源域和目标域之间的分布,或学习一个在所有领域中都表现良好的不变特征空间。然而,这两种策略都需要在训练过程中访问测试域的足够数量的样本,尽管不一定需要注释。这一限制阻碍了模型在临床环境中的广泛部署,因为目标域数据可能只能在实时环境中访问。在这项工作中,我们引入了HyDA,这是一种新型超网络框架,它利用领域特性而不是抑制它们,从而在推理时间实现动态适应。具体来说,HyDA学习隐式领域表示,并使用它们在运行时实时调整模型参数,有效地插值到未见过的领域。我们在两个临床相关的应用上验证了HyDA的有效性,分别是MRI脑龄预测和胸部X射线病理分类,展示了它在跨任务和跨模态方面的泛化能力。我们的代码将在(待定)处提供。
论文及项目相关链接
PDF submitted to MICCAI 2025
Summary
医学成像数据集由于采集协议、患者特征和成像设备差异而常有变化。这些被称为域偏移的数据分布变化,为将成像分析模型适应于实际医疗应用带来了挑战。当前大多数域适应方法旨在对齐源域和目标域分布,或学习一个在所有域中表现良好的不变特征空间。然而,这两种策略都需要在训练期间访问测试域的足够样本,即使不必进行标注。这一局限性阻碍了模型在临床环境中的广泛应用,因为在现实时间中可能仅可访问目标域数据。在此工作中,我们引入了HyDA,这是一种利用域特征而非抑制它们的超网络框架,可在推断时进行动态适应。具体而言,HyDA学习隐式域表示并使用它们即时调整模型参数,有效地推广到未见过的域。我们在两个临床应用上验证了HyDA的有效性,分别是MRI脑部年龄预测和胸部X射线病理分类,证明了其跨任务和模态的泛化能力。
Key Takeaways
- 医学成像数据集存在由于采集协议、患者特征和成像设备差异导致的域偏移问题。
- 当前域适应方法主要通过对齐域分布或学习不变特征空间来适应模型,但需访问测试域的样本,这在临床环境中存在局限性。
- HyDA是一种新型超网络框架,利用隐式域表示进行动态适应,无需在训练期间访问目标域样本。
- HyDA在MRI脑部年龄预测和胸部X射线病理分类等临床应用中得到了验证。
- HyDA具有跨任务和模态的泛化能力。
- 该方法实现了动态参数调整以适应不同域的样本,展现出对未见域的泛化潜力。
点此查看论文截图

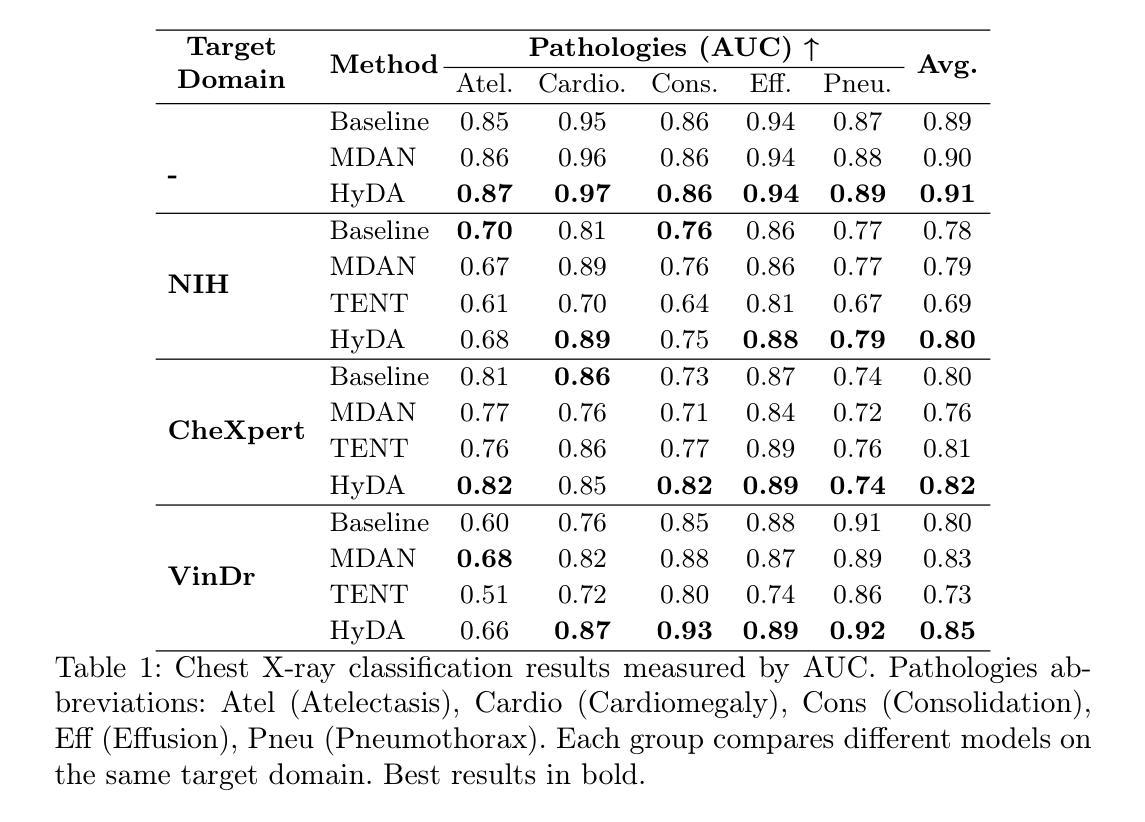
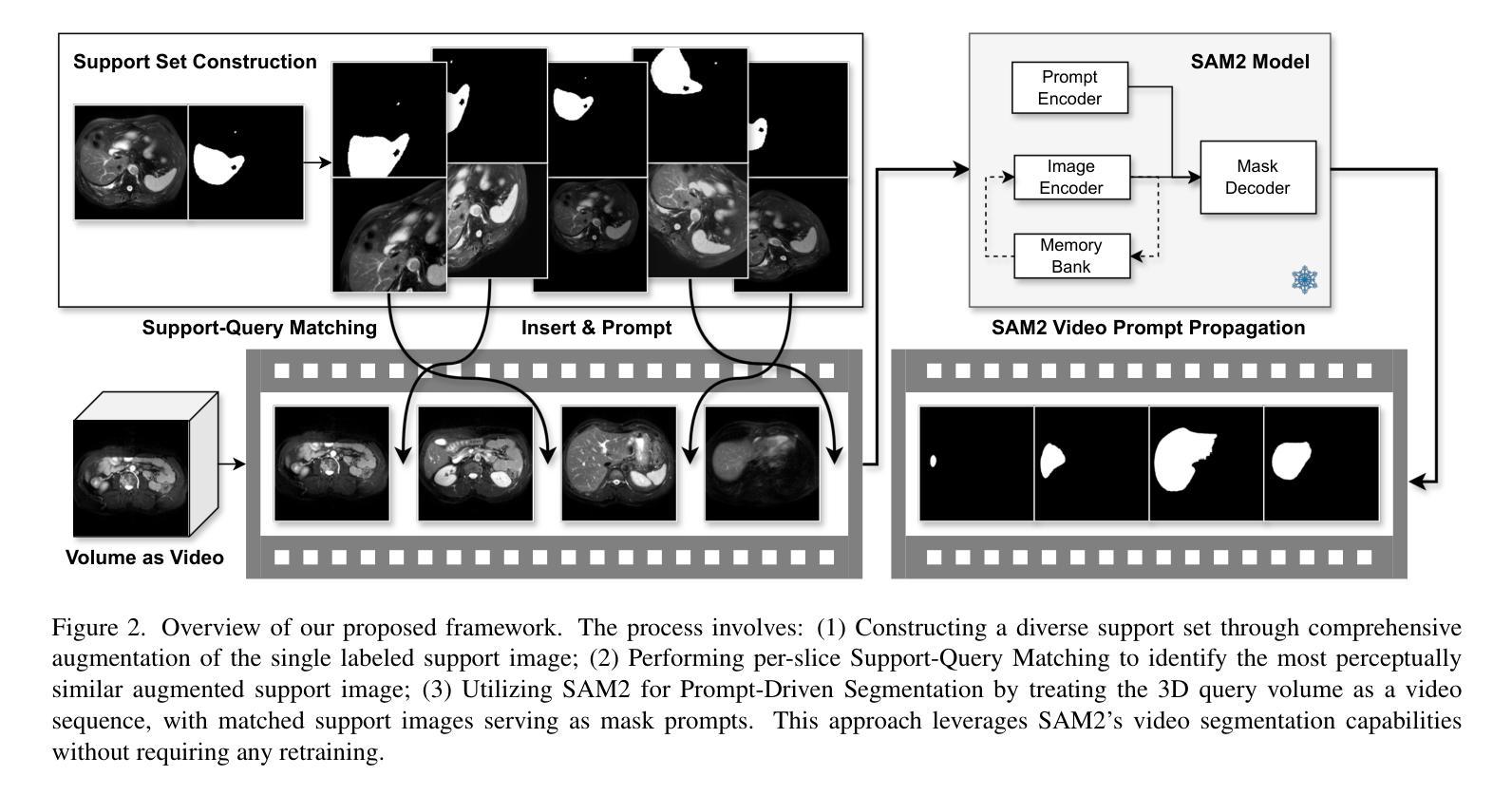
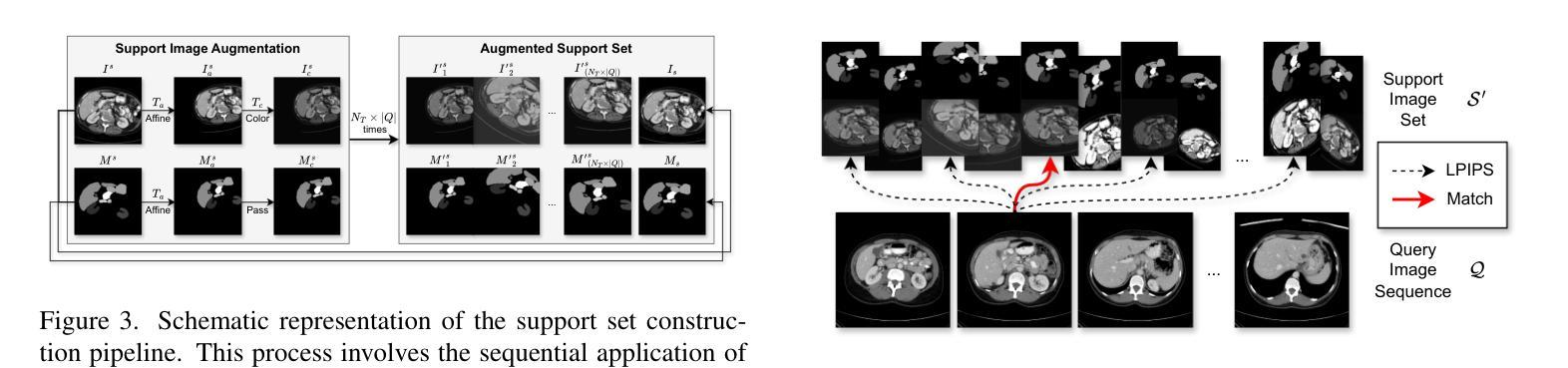
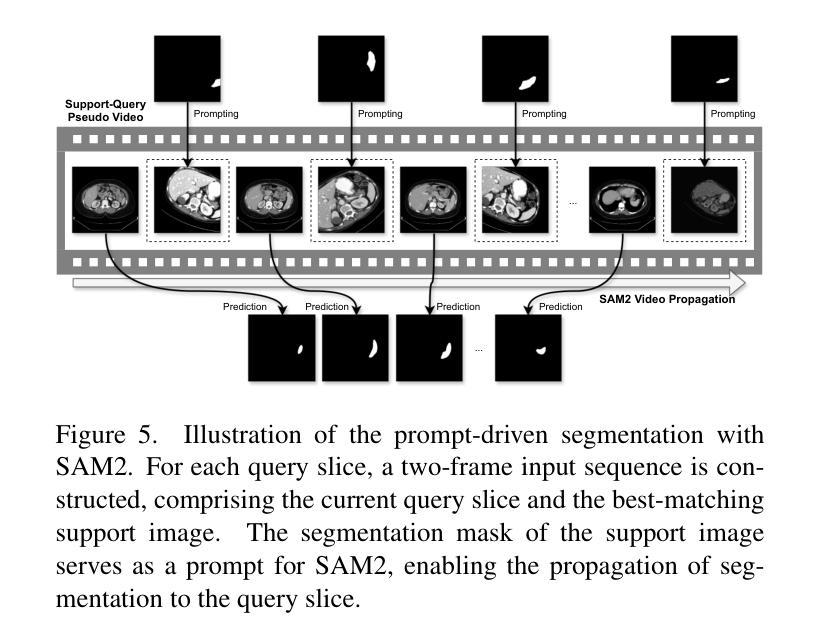
Rethinking Few-Shot Medical Image Segmentation by SAM2: A Training-Free Framework with Augmentative Prompting and Dynamic Matching
Authors:Haiyue Zu, Jun Ge, Heting Xiao, Jile Xie, Zhangzhe Zhou, Yifan Meng, Jiayi Ni, Junjie Niu, Linlin Zhang, Li Ni, Huilin Yang
The reliance on large labeled datasets presents a significant challenge in medical image segmentation. Few-shot learning offers a potential solution, but existing methods often still require substantial training data. This paper proposes a novel approach that leverages the Segment Anything Model 2 (SAM2), a vision foundation model with strong video segmentation capabilities. We conceptualize 3D medical image volumes as video sequences, departing from the traditional slice-by-slice paradigm. Our core innovation is a support-query matching strategy: we perform extensive data augmentation on a single labeled support image and, for each frame in the query volume, algorithmically select the most analogous augmented support image. This selected image, along with its corresponding mask, is used as a mask prompt, driving SAM2’s video segmentation. This approach entirely avoids model retraining or parameter updates. We demonstrate state-of-the-art performance on benchmark few-shot medical image segmentation datasets, achieving significant improvements in accuracy and annotation efficiency. This plug-and-play method offers a powerful and generalizable solution for 3D medical image segmentation.
在医学图像分割中,对大标签数据集的依赖构成了一大挑战。小样本学习提供了一个潜在的解决方案,但现有方法往往仍需要大量训练数据。本文提出了一种利用Segment Anything Model 2(SAM2)的新方法,这是一个具有强大视频分割能力的视觉基础模型。我们将3D医学图像体积概念化为视频序列,摒弃了传统的逐片处理模式。我们的核心创新之处在于支持查询匹配策略:我们对单个标记的支持图像进行大量数据增强,并针对查询体积中的每一帧,算法选择最类似的增强支持图像。所选图像及其相应的掩膜被用作掩膜提示,驱动SAM2的视频分割。此方法完全避免了模型重新训练或参数更新。我们在基准小样本医学图像分割数据集上展示了最先进的性能,在准确性和注释效率方面取得了显著改进。这种即插即用方法为3D医学图像分割提供了强大且通用的解决方案。
论文及项目相关链接
Summary
本论文提出了一种新的医学图像分割方法,利用Segment Anything Model 2(SAM2)模型强大的视频分割能力。通过将3D医学图像体积概念化为视频序列,采用支持查询匹配策略,对单个标记的支持图像进行大量数据增强,并算法选择最相似的增强支持图像为查询体积的每一帧进行分割。此方法避免了模型重新训练或参数更新,提高了准确性和标注效率,为三维医学图像分割提供了强大且通用的解决方案。
Key Takeaways
- 医学图像分割中依赖大量标记数据集是一大挑战。
- Few-shot学习提供了一种潜在的解决方案,但现有方法仍需要大量训练数据。
- 本论文提出一种新型方法,利用Segment Anything Model 2(SAM2)模型进行医学图像分割。
- 将3D医学图像体积概念化为视频序列,采用支持查询匹配策略进行数据增强。
- 该方法通过选择最相似的增强支持图像为查询体积的每一帧进行分割,避免了模型重新训练或参数更新。
- 在基准少样本医学图像分割数据集上实现了最先进的性能,显著提高了准确性和标注效率。
点此查看论文截图

Spatial regularisation for improved accuracy and interpretability in keypoint-based registration
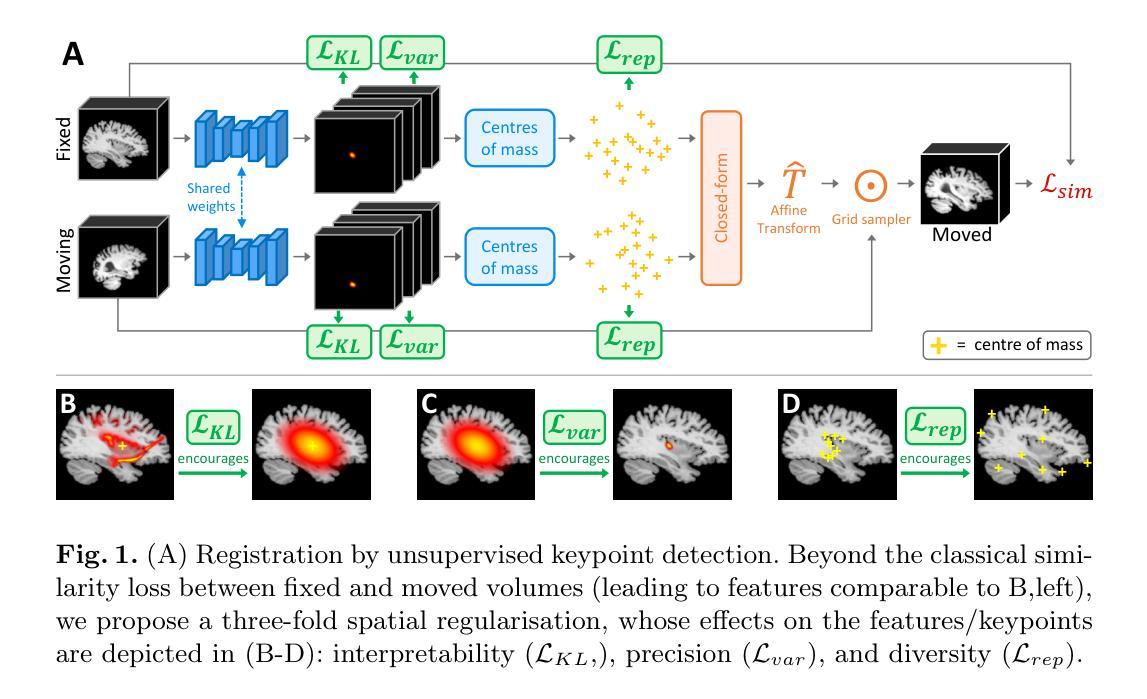
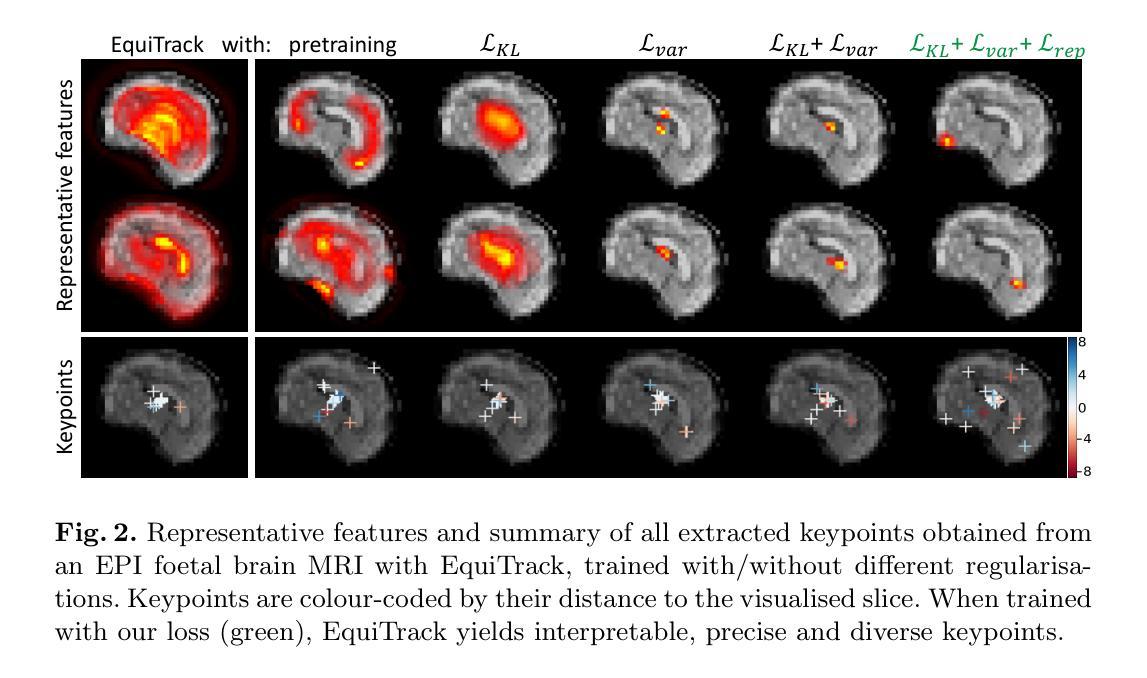
Authors:Benjamin Billot, Ramya Muthukrishnan, Esra Abaci-Turk, P. Ellen Grant, Nicholas Ayache, Hervé Delingette, Polina Golland
Unsupervised registration strategies bypass requirements in ground truth transforms or segmentations by optimising similarity metrics between fixed and moved volumes. Among these methods, a recent subclass of approaches based on unsupervised keypoint detection stand out as very promising for interpretability. Specifically, these methods train a network to predict feature maps for fixed and moving images, from which explainable centres of mass are computed to obtain point clouds, that are then aligned in closed-form. However, the features returned by the network often yield spatially diffuse patterns that are hard to interpret, thus undermining the purpose of keypoint-based registration. Here, we propose a three-fold loss to regularise the spatial distribution of the features. First, we use the KL divergence to model features as point spread functions that we interpret as probabilistic keypoints. Then, we sharpen the spatial distributions of these features to increase the precision of the detected landmarks. Finally, we introduce a new repulsive loss across keypoints to encourage spatial diversity. Overall, our loss considerably improves the interpretability of the features, which now correspond to precise and anatomically meaningful landmarks. We demonstrate our three-fold loss in foetal rigid motion tracking and brain MRI affine registration tasks, where it not only outperforms state-of-the-art unsupervised strategies, but also bridges the gap with state-of-the-art supervised methods. Our code is available at https://github.com/BenBillot/spatial_regularisation.
无监督的注册策略通过优化固定体积和移动体积之间的相似度指标,从而绕过了对真实变换或分割的要求。在这些方法中,最近出现的一种基于无监督关键点检测的方法作为解释性很强的子类脱颖而出。具体来说,这些方法训练网络对固定图像和移动图像进行特征映射预测,从中计算出可解释的质量中心,以获得点云,然后进行闭式对齐。然而,网络返回的特征通常会产生难以解释的空间扩散模式,从而破坏了基于关键点的注册目的。在这里,我们提出了一个三重损失来规范特征的空间分布。首先,我们使用KL散度将特征建模为点扩散函数,我们将其解释为概率关键点。然后,我们锐化这些特征的空间分布,以提高检测到的地标的精度。最后,我们在关键点之间引入了一个新的排斥损失来鼓励空间多样性。总的来说,我们的损失大大提高了特征的解释性,这些特征现在对应于精确且解剖上意义重大的地标。我们在胎儿刚体运动跟踪和脑部MRI仿射注册任务中展示了我们的三重损失,它不仅超越了最先进的无监督策略,而且缩小了与最先进的监督方法的差距。我们的代码可在https://github.com/BenBillot/spatial_regularisation访问。
论文及项目相关链接
PDF under review
Summary
本文介绍了一种基于无监督关键点检测的方法,用于医学图像配准。该方法通过优化固定图像和移动图像之间的相似性度量来实现无监督配准。针对网络特征返回的空间扩散模式难以解释的问题,提出了三倍损失来规范特征的空间分布。通过KL散度建模特征为点扩散函数,尖锐化特征的空间分布,并引入排斥损失鼓励空间多样性。改进后的方法在胎儿刚性运动跟踪和MRI脑部图像仿射配准任务中表现优异,其性能超越了现有无监督策略,并接近了监督方法的水平。
Key Takeaways
- 无监督配准策略通过优化固定和移动图像之间的相似性度量,绕过对真实标签变换或分割的要求。
- 基于无监督关键点检测的方法在医学图像配准中展现出巨大潜力。
- 网络特征返回的空间扩散模式难以解释,影响关键点配准的效果。
- 提出了一种三倍损失来规范特征的空间分布,包括使用KL散度建模特征、尖锐化特征空间分布以及引入排斥损失。
- 改进后的方法在胎儿刚性运动跟踪和MRI脑部图像仿射配准任务中表现优异。
- 该方法不仅超越了现有无监督策略,还缩小了与现有监督方法的差距。
点此查看论文截图

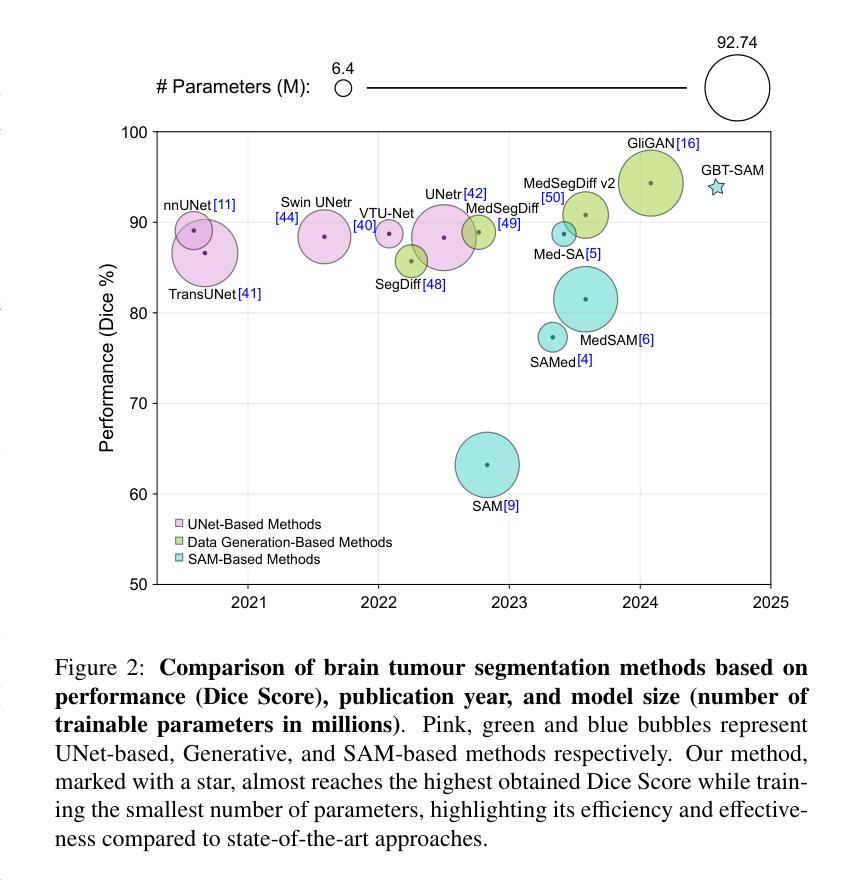
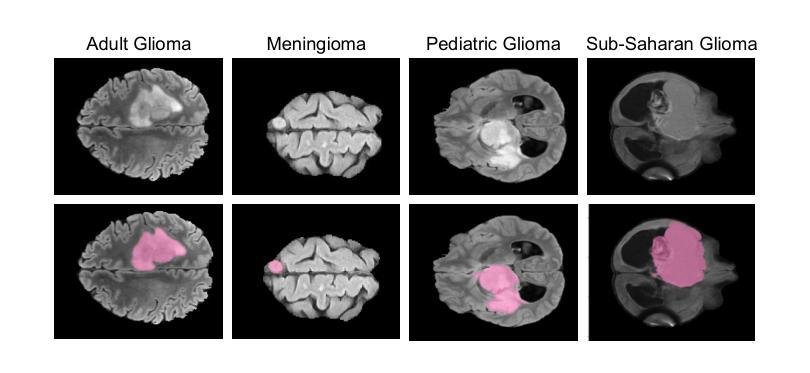
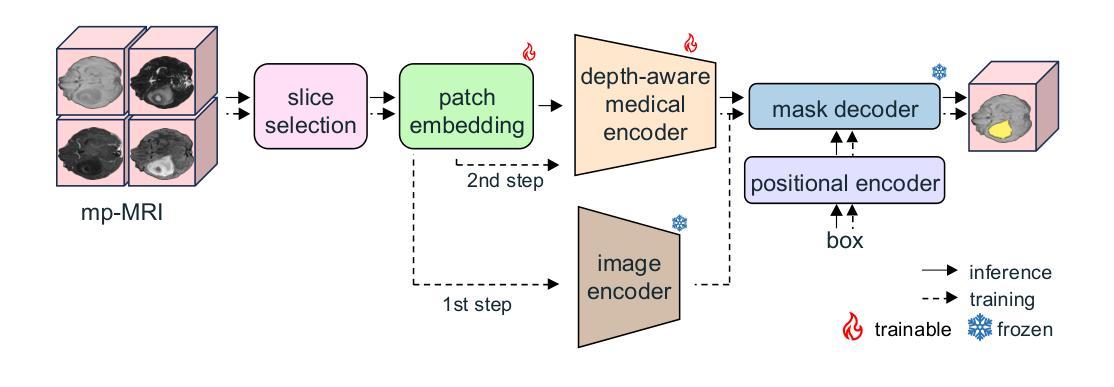
GBT-SAM: A Parameter-Efficient Depth-Aware Model for Generalizable Brain tumour Segmentation on mp-MRI
Authors:Cecilia Diana-Albelda, Roberto Alcover-Couso, Álvaro García-Martín, Jesus Bescos, Marcos Escudero-Viñolo
Gliomas are brain tumours that stand out for their highly lethal and aggressive nature, which demands a precise approach in their diagnosis. Medical image segmentation plays a crucial role in the evaluation and follow-up of these tumours, allowing specialists to analyse their morphology. However, existing methods for automatic glioma segmentation often lack generalization capability across other brain tumour domains, require extensive computational resources, or fail to fully utilize the multi-parametric MRI (mp-MRI) data used to delineate them. In this work, we introduce GBT-SAM, a novel Generalizable Brain Tumour (GBT) framework that extends the Segment Anything Model (SAM) to brain tumour segmentation tasks. Our method employs a two-step training protocol: first, fine-tuning the patch embedding layer to process the entire mp-MRI modalities, and second, incorporating parameter-efficient LoRA blocks and a Depth-Condition block into the Vision Transformer (ViT) to capture inter-slice correlations. GBT-SAM achieves state-of-the-art performance on the Adult Glioma dataset (Dice Score of $93.54$) while demonstrating robust generalization across Meningioma, Pediatric Glioma, and Sub-Saharan Glioma datasets. Furthermore, GBT-SAM uses less than 6.5M trainable parameters, thus offering an efficient solution for brain tumour segmentation. \ Our code and models are available at https://github.com/vpulab/med-sam-brain .
胶质瘤是突出的脑肿瘤,具有极高的致死性和侵袭性,这要求在诊断时采取精确的方法。医学图像分割在评估和跟踪这些肿瘤中起着至关重要的作用,允许专家分析它们的形态。然而,现有的自动胶质瘤分割方法往往缺乏在其他脑肿瘤领域的泛化能力,需要大量的计算资源,或者未能充分利用用于描绘它们的多参数MRI(mp-MRI)数据。在这项工作中,我们介绍了GBT-SAM,这是一个新的通用脑肿瘤(GBT)框架,它将Segment Anything Model(SAM)扩展到脑肿瘤分割任务。我们的方法采用两步训练协议:首先,微调补丁嵌入层以处理整个mp-MRI模式;其次,在视觉变压器(ViT)中融入参数高效的LoRA块和深度条件块,以捕捉跨切片的相关性。GBT-SAM在成人胶质瘤数据集上实现了最先进的性能(Dice得分为93.54%),同时显示出在脑膜瘤、儿童胶质瘤和撒哈拉以南胶质瘤数据集上的稳健泛化。此外,GBT-SAM使用的可训练参数少于650万,因此为脑肿瘤分割提供了有效的解决方案。我们的代码和模型可在https://github.com/vpulab/med-sam-brain上找到。
论文及项目相关链接
Summary
本文介绍了一种新型通用脑肿瘤(GBT)分割框架GBT-SAM,它扩展了Segment Anything Model(SAM)以进行脑肿瘤分割任务。该方法采用两步训练协议,首先微调补丁嵌入层以处理整个多参数MRI(mp-MRI)模式,其次在视觉转换器(ViT)中融入参数高效的LoRA块和深度条件块以捕获切片间的相关性。GBT-SAM在成人胶质瘤数据集上取得了最先进的性能(Dice得分为93.54%),并在脑膜瘤、儿童胶质瘤和撒哈拉以南胶质瘤数据集上表现出稳健的泛化能力。此外,GBT-SAM使用的可训练参数少于650万,为脑肿瘤分割提供了有效的解决方案。
Key Takeaways
- GBT-SAM是一种针对脑肿瘤分割的新型框架,基于Segment Anything Model(SAM)。
- 该方法采用两步训练协议,包括微调补丁嵌入层和融入参数高效的LoRA块及深度条件块。
- GBT-SAM在成人胶质瘤数据集上取得高Dice得分,并展示了对不同类型脑肿瘤的稳健泛化能力。
- GBT-SAM使用的可训练参数较少,为脑肿瘤分割提供了高效解决方案。
- GBT-SAM方法充分利用了多参数MRI数据,提高了脑肿瘤分割的准确性和效率。
点此查看论文截图

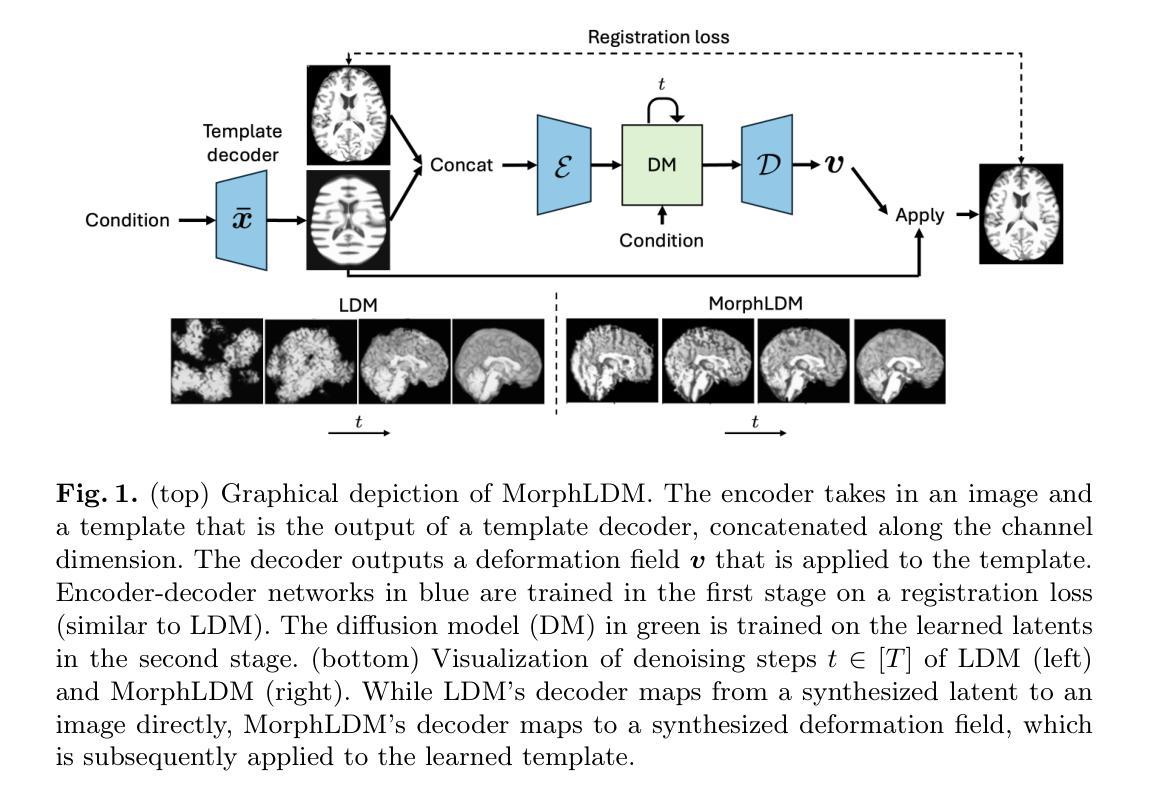
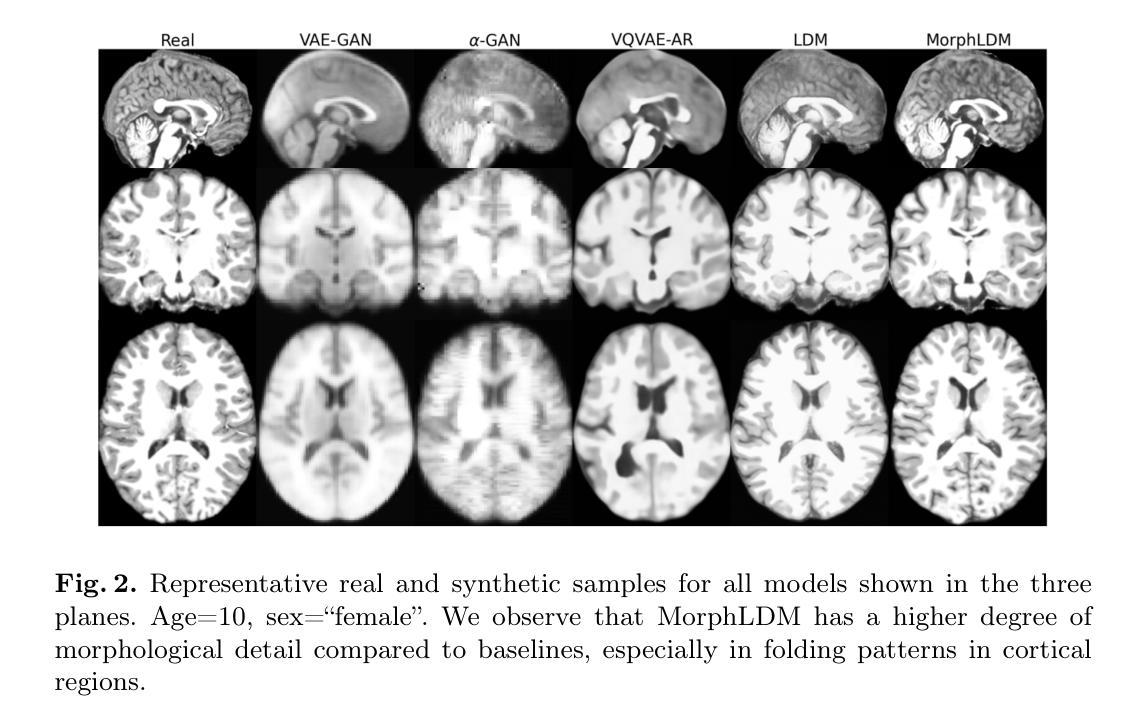
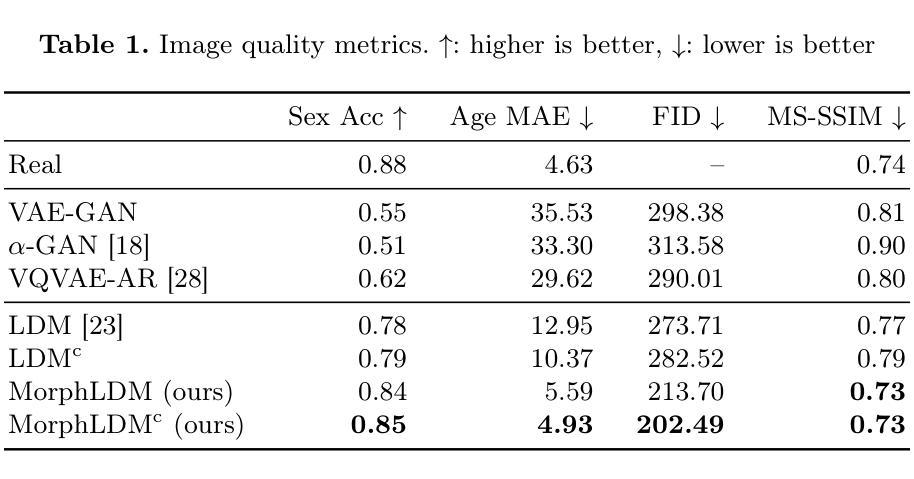
Generating Novel Brain Morphology by Deforming Learned Templates
Authors:Alan Q. Wang, Fangrui Huang, Bailey Trang, Wei Peng, Mohammad Abbasi, Kilian Pohl, Mert Sabuncu, Ehsan Adeli
Designing generative models for 3D structural brain MRI that synthesize morphologically-plausible and attribute-specific (e.g., age, sex, disease state) samples is an active area of research. Existing approaches based on frameworks like GANs or diffusion models synthesize the image directly, which may limit their ability to capture intricate morphological details. In this work, we propose a 3D brain MRI generation method based on state-of-the-art latent diffusion models (LDMs), called MorphLDM, that generates novel images by applying synthesized deformation fields to a learned template. Instead of using a reconstruction-based autoencoder (as in a typical LDM), our encoder outputs a latent embedding derived from both an image and a learned template that is itself the output of a template decoder; this latent is passed to a deformation field decoder, whose output is applied to the learned template. A registration loss is minimized between the original image and the deformed template with respect to the encoder and both decoders. Empirically, our approach outperforms generative baselines on metrics spanning image diversity, adherence with respect to input conditions, and voxel-based morphometry. Our code is available at https://github.com/alanqrwang/morphldm.
设计用于合成形态上合理且具备特定属性(例如年龄、性别、疾病状态)样本的3D结构脑MRI生成模型是一个热门研究领域。现有基于GAN或扩散模型等框架的方法直接合成图像,这可能限制了它们在捕捉复杂形态细节方面的能力。在这项工作中,我们提出了一种基于最新潜在扩散模型(LDM)的3D脑MRI生成方法,称为MorphLDM。该方法通过应用合成变形场于已学习模板来生成新型图像。与其他使用基于重建的自编码器(如典型LDM中的)不同,我们的编码器输出一个潜在嵌入,该嵌入来源于图像和已学习模板,而该模板本身也是模板解码器的输出;此潜在嵌入被传递给变形场解码器,其输出应用于已学习模板。通过最小化原始图像和变形模板之间的注册损失,关于编码器和两个解码器的损失都会被优化。从实证结果来看,我们的方法在图像多样性、符合输入条件以及基于体素的形态测量等指标上的表现均超过了基准生成模型。我们的代码可在https://github.com/alanqrwang/morphldm找到。
论文及项目相关链接
Summary
基于最新潜在扩散模型(LDM)的3D脑MRI生成方法MorphLDM,通过应用合成变形场于学习模板以生成新图像。相较于传统LDM使用重建自编码器,MorphLDM的编码器输出一个由图像和学习模板共同得出的潜在嵌入,然后传递给变形场解码器,其输出应用于学习模板。通过最小化原始图像与变形模板之间的注册损失,以提高图像多样性、符合输入条件以及基于体素的形态测量等指标。
Key Takeaways
- 研究领域:该研究关注于设计用于3D结构脑MRI的生成模型,这些模型能够合成形态上合理且具备特定属性(如年龄、性别、疾病状态)的样本。
- 方法创新:提出了一种基于最新潜在扩散模型(LDM)的3D脑MRI生成方法MorphLDM。不同于传统直接合成图像的方法,MorphLDM通过应用合成变形场到学习模板来生成新图像。
- 编码器的特点:MorphLDM的编码器输出一个由图像和学习模板共同产生的潜在嵌入。学习模板本身是模板解码器的输出。
- 变形场和注册损失:通过最小化原始图像和变形模板之间的注册损失,以提高图像合成的质量。这一损失涉及到编码器和两个解码器。
- 性能评估:该方法的性能通过一系列指标进行评估,包括图像多样性、符合输入条件的能力以及基于体素的形态测量。实证结果表明,该方法优于其他生成方法。
- 代码共享:研究团队已将相关代码上传至GitHub(链接:https://github.com/alanqrwang/morphldm)。
点此查看论文截图

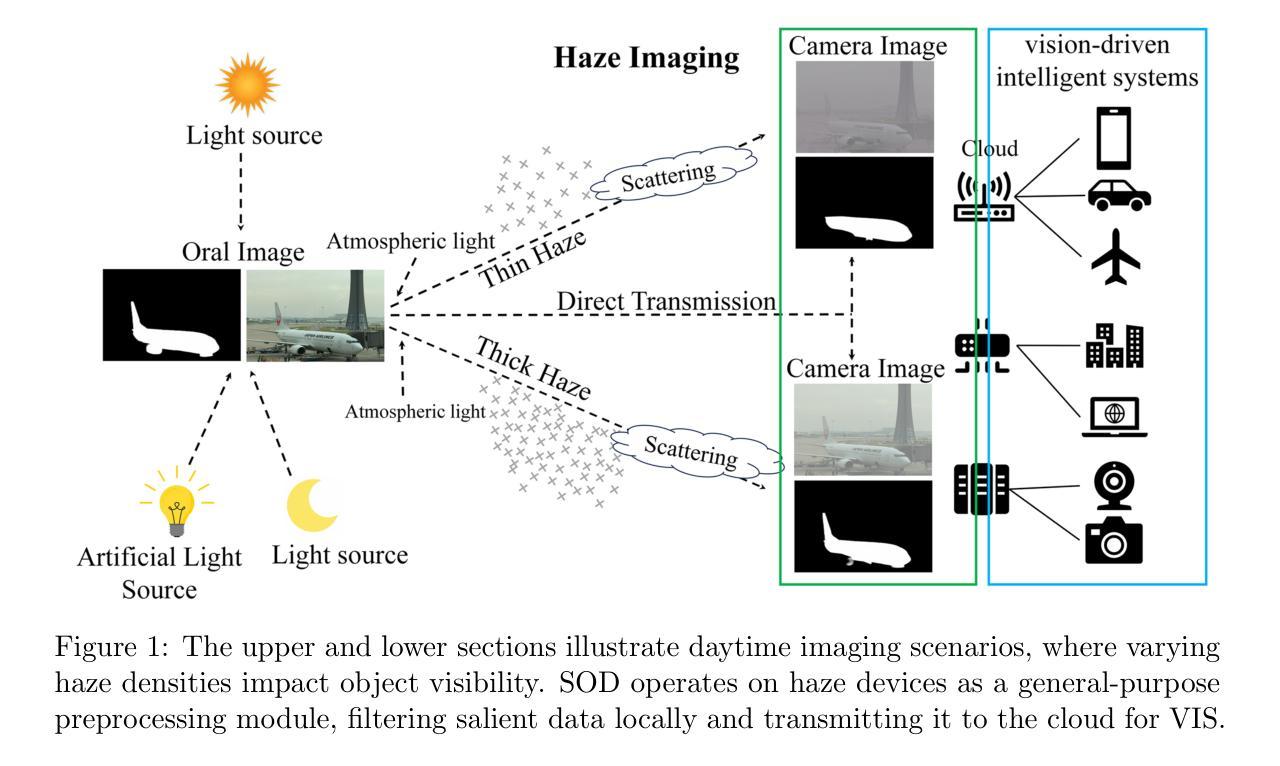
Multi-Knowledge-oriented Nighttime Haze Imaging Enhancer for Vision-driven Intelligent Systems
Authors:Ai Chen, Yuxu Lu, Dong Yang, Junlin Zhou, Yan Fu, Duanbing Chen
Salient object detection (SOD) plays a critical role in vision-driven measurement systems (VMS), facilitating the detection and segmentation of key visual elements in an image. However, adverse imaging conditions such as haze during the day, low light, and haze at night severely degrade image quality, and complicating the SOD process. To address these challenges, we propose a multi-task-oriented nighttime haze imaging enhancer (MToIE), which integrates three tasks: daytime dehazing, low-light enhancement, and nighttime dehazing. The MToIE incorporates two key innovative components: First, the network employs a task-oriented node learning mechanism to handle three specific degradation types: day-time haze, low light, and night-time haze conditions, with an embedded self-attention module enhancing its performance in nighttime imaging. In addition, multi-receptive field enhancement module that efficiently extracts multi-scale features through three parallel depthwise separable convolution branches with different dilation rates, capturing comprehensive spatial information with minimal computational overhead. To ensure optimal image reconstruction quality and visual characteristics, we suggest a hybrid loss function. Extensive experiments on different types of weather/imaging conditions illustrate that MToIE surpasses existing methods, significantly enhancing the accuracy and reliability of vision systems across diverse imaging scenarios. The code is available at https://github.com/Ai-Chen-Lab/MKoIE.
显著性目标检测(SOD)在视觉驱动测量系统(VMS)中扮演着至关重要的角色,它促进了图像中关键视觉元素的检测和分割。然而,不利的成像条件,如白天的雾霾、低光照和夜晚的雾霾,严重降低了图像质量,并使得SOD过程复杂化。为了应对这些挑战,我们提出了一种多任务的夜间雾霾成像增强器(MToIE),它结合了三项任务:白天去雾、低光增强和夜间去雾。MToIE包含两个关键的创新组件:首先,网络采用任务导向的节点学习机制来处理三种特定的退化类型:白天雾霾、低光和夜间雾霾条件,其中嵌入的自注意模块增强了其在夜间成像的性能。此外,多感受野增强模块通过三个并行深度可分离卷积分支有效地提取多尺度特征,这些分支具有不同的膨胀率,以最小的计算开销捕获全面的空间信息。为了确保最佳的图像重建质量和视觉特性,我们提出了一种混合损失函数。在不同天气/成像条件下的大量实验表明,MToIE超越了现有方法,显著提高了不同成像场景中视觉系统的准确性和可靠性。代码可在https://github.com/Ai-Chen-Lab/MKoIE找到。
论文及项目相关链接
Summary
针对日间雾霾、低光照和夜间雾霾等恶劣成像条件对视觉系统的影响,本文提出了一种多任务导向的夜间雾霾成像增强器(MToIE)。它通过集成日间去雾、低光增强和夜间去雾三项任务,采用任务导向节点学习机制和自注意力模块,并结合多感受野增强模块和混合损失函数,提高了成像质量,显著提升了视觉系统在多种成像场景下的准确性和可靠性。
Key Takeaways
- Salient object detection (SOD)在恶劣成像条件下(如日间雾霾、低光照和夜间雾霾)面临挑战。
- 提出了一种多任务导向的夜间雾霾成像增强器(MToIE),集成了三项任务:日间去雾、低光增强和夜间去雾。
- MToIE采用任务导向节点学习机制和自注意力模块,以处理不同类型的恶劣成像条件并增强夜间成像性能。
- MToIE引入了多感受野增强模块,通过三个具有不同膨胀率的并行深度可分离卷积分支来有效地提取多尺度特征,从而捕获全面的空间信息并保持较低的计算开销。
- 为了确保图像重建质量和视觉特性,建议使用混合损失函数。
- 广泛的实验表明,MToIE在多种天气/成像条件下超越了现有方法,显著提高了视觉系统的准确性和可靠性。
- 代码已公开在GitHub上(https://github.com/Ai-Chen-Lab/MKoIE)。
点此查看论文截图

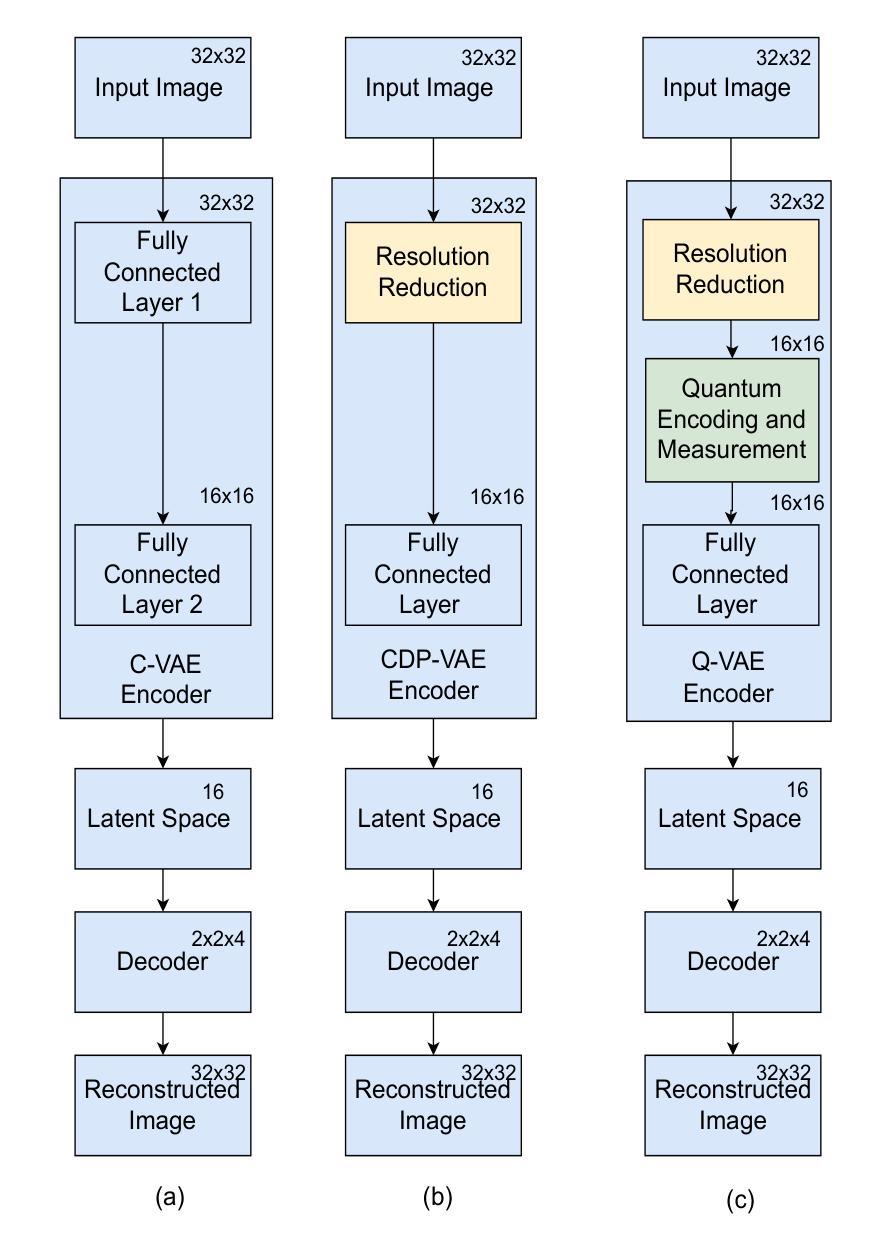
Quantum Down Sampling Filter for Variational Auto-encoder
Authors:Farina Riaz, Fakhar Zaman, Hajime Suzuki, Sharif Abuadbba, David Nguyen
Variational autoencoders (VAEs) are fundamental for generative modeling and image reconstruction, yet their performance often struggles to maintain high fidelity in reconstructions. This study introduces a hybrid model, quantum variational autoencoder (Q-VAE), which integrates quantum encoding within the encoder while utilizing fully connected layers to extract meaningful representations. The decoder uses transposed convolution layers for up-sampling. The Q-VAE is evaluated against the classical VAE and the classical direct-passing VAE, which utilizes windowed pooling filters. Results on the MNIST and USPS datasets demonstrate that Q-VAE consistently outperforms classical approaches, achieving lower Fr'echet inception distance scores, thereby indicating superior image fidelity and enhanced reconstruction quality. These findings highlight the potential of Q-VAE for high-quality synthetic data generation and improved image reconstruction in generative models.
变分自编码器(VAEs)在生成建模和图像重建中具有重要作用,但其性能在保持高保真重建方面往往存在挑战。本研究引入了一种混合模型,即量子变分自编码器(Q-VAE),它在编码器内集成了量子编码,同时使用全连接层提取有意义的表示。解码器使用转置卷积层进行上采样。Q-VAE与经典VAE和经典直通VAE(使用窗口池化滤波器)进行了评估比较。在MNIST和USPS数据集上的结果表明,Q-VAE始终优于经典方法,实现了更低的Fréchet inception距离得分,从而表明其图像保真度更高,重建质量增强。这些发现突出了Q-VAE在高质量合成数据生成和生成模型中改进图像重建的潜力。
论文及项目相关链接
PDF 18 pages, 13 figures
Summary
本文介绍了量子变分自编码器(Q-VAE)的研究,这是一种结合了量子编码技术的混合模型。相较于传统的变分自编码器(VAE)和直接传递的VAE,Q-VAE在提取有意义表示方面表现更佳,使用了转置卷积层进行上采样。在MNIST和USPS数据集上的实验结果表明,Q-VAE具有更低的Fréchet inception距离得分,表明其图像保真度和重建质量更高,展现出其在高质量合成数据生成和改进生成模型中的图像重建潜力。
Key Takeaways
- 量子变分自编码器(Q-VAE)结合了量子编码技术与变分自编码器(VAE)。
- Q-VAE使用全连接层提取有意义的表示。
- 解码器采用转置卷积层进行上采样。
- 在MNIST和USPS数据集上,Q-VAE相较于传统方法表现出更高的性能。
- Q-VAE具有更低的Fréchet inception距离得分,表明其图像重建质量更高。
- Q-VAE具有潜力用于高质量合成数据生成。
点此查看论文截图