⚠️ 以下所有内容总结都来自于 大语言模型的能力,如有错误,仅供参考,谨慎使用
🔴 请注意:千万不要用于严肃的学术场景,只能用于论文阅读前的初筛!
💗 如果您觉得我们的项目对您有帮助 ChatPaperFree ,还请您给我们一些鼓励!⭐️ HuggingFace免费体验
2025-03-12 更新
LLaVA-RadZ: Can Multimodal Large Language Models Effectively Tackle Zero-shot Radiology Recognition?
Authors:Bangyan Li, Wenxuan Huang, Yunhang Shen, Yeqiang Wang, Shaohui Lin, Jingzhong Lin, Ling You, Yinqi Zhang, Ke Li, Xing Sun, Yuling Sun
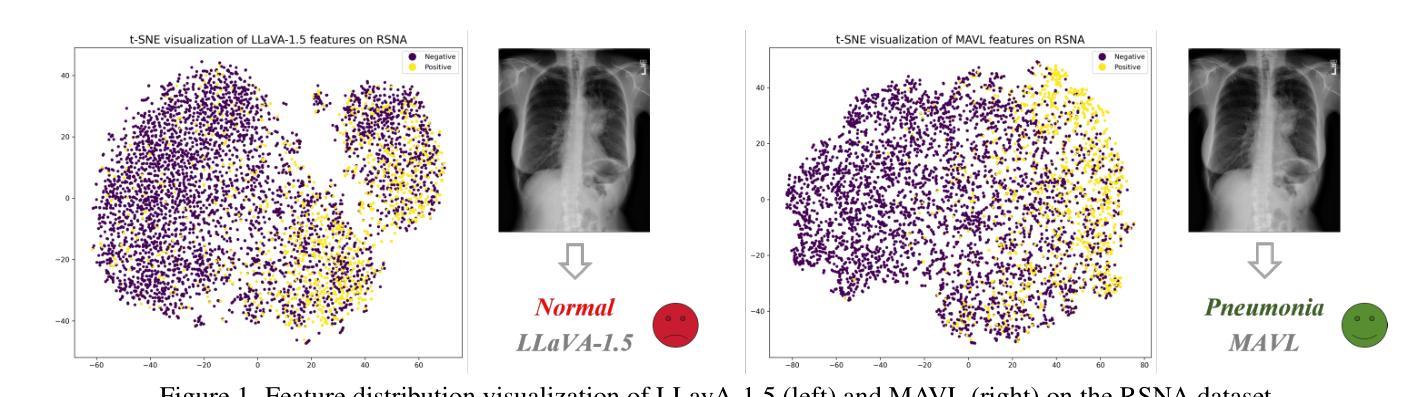
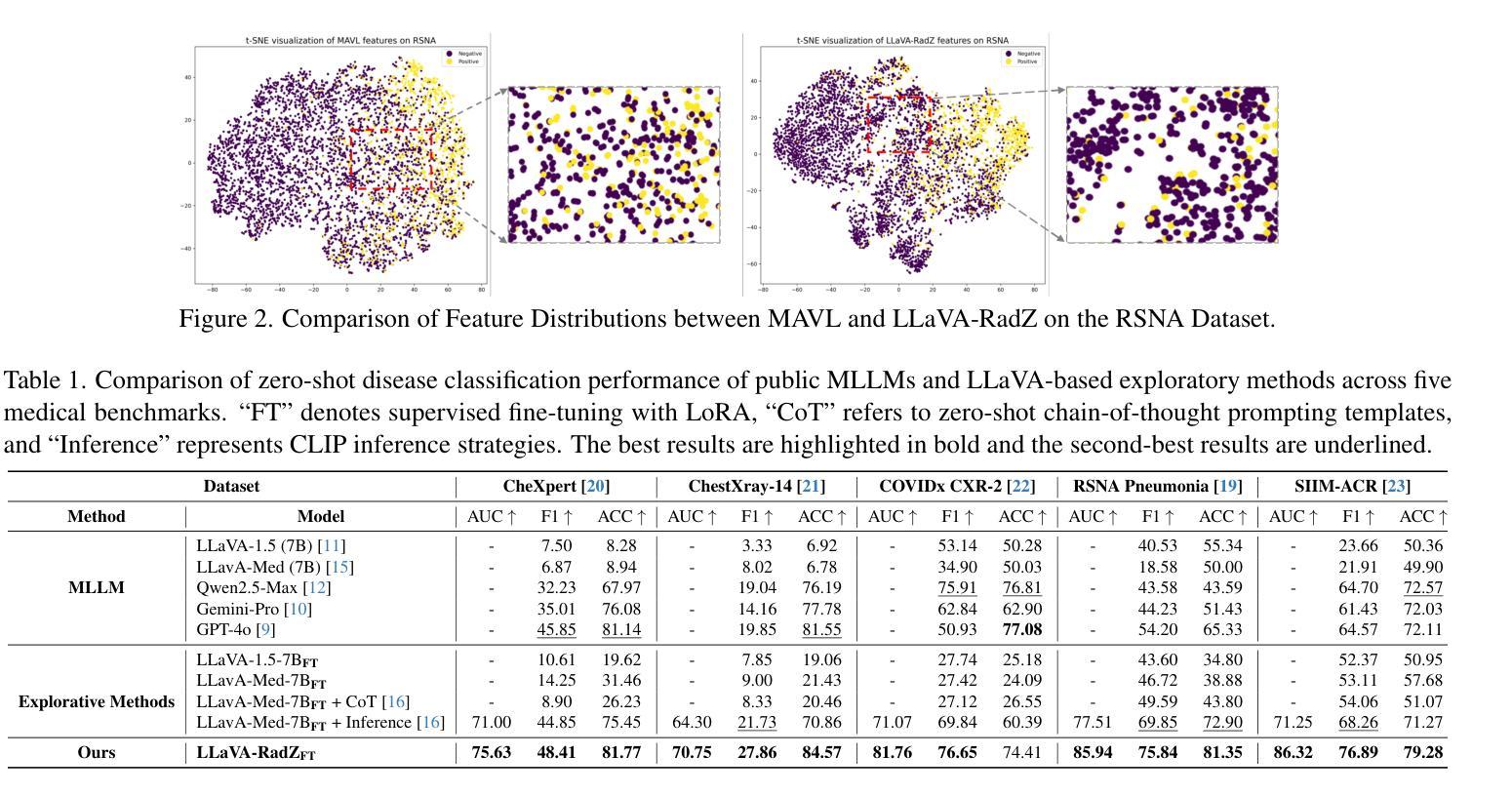
Recently, multimodal large models (MLLMs) have demonstrated exceptional capabilities in visual understanding and reasoning across various vision-language tasks. However, MLLMs usually perform poorly in zero-shot medical disease recognition, as they do not fully exploit the captured features and available medical knowledge. To address this challenge, we propose LLaVA-RadZ, a simple yet effective framework for zero-shot medical disease recognition. Specifically, we design an end-to-end training strategy, termed Decoding-Side Feature Alignment Training (DFAT) to take advantage of the characteristics of the MLLM decoder architecture and incorporate modality-specific tokens tailored for different modalities, which effectively utilizes image and text representations and facilitates robust cross-modal alignment. Additionally, we introduce a Domain Knowledge Anchoring Module (DKAM) to exploit the intrinsic medical knowledge of large models, which mitigates the category semantic gap in image-text alignment. DKAM improves category-level alignment, allowing for accurate disease recognition. Extensive experiments on multiple benchmarks demonstrate that our LLaVA-RadZ significantly outperforms traditional MLLMs in zero-shot disease recognition and exhibits the state-of-the-art performance compared to the well-established and highly-optimized CLIP-based approaches.
最近,多模态大型模型(MLLMs)在各种视觉语言任务中表现出了出色的视觉理解和推理能力。然而,在零样本医疗疾病识别方面,MLLMs的表现通常不佳,因为它们没有完全挖掘出捕获的特征和可用的医学知识。为了解决这一挑战,我们提出了LLaVA-RadZ,这是一个用于零样本医疗疾病识别的简单有效的框架。具体来说,我们设计了一种端到端的训练策略,称为解码侧特征对齐训练(DFAT),以利用MLLM解码器的特点,并结合针对不同模态定制的模态特定令牌,这有效地利用了图像和文本表示,并促进了稳健的跨模态对齐。此外,我们引入了领域知识锚定模块(DKAM),以利用大型模型的内在医学知识,这减轻了图像文本对齐中的类别语义鸿沟。DKAM改善了类别级别的对齐,从而实现准确的疾病识别。在多个基准测试上的广泛实验表明,我们的LLaVA-RadZ在零样本疾病识别方面显著优于传统的MLLMs,与建立良好且高度优化的CLIP方法相比,具有最先进的性能。
论文及项目相关链接
Summary
针对多模态大型模型在零样本医疗疾病识别中的不足,提出LLaVA-RadZ框架,通过解码侧特征对齐训练(DFAT)和领域知识锚定模块(DKAM),有效融合图像和文本表征,实现稳健的跨模态对齐和疾病识别。在多个基准测试上表现优异,显著优于传统MLLMs和高度优化的CLIP方法。
Key Takeaways
- 多模态大型模型(MLLMs)在视觉理解和推理方面表现出卓越的能力,但在零样本医疗疾病识别中存在挑战。
- LLaVA-RadZ框架旨在解决MLLMs在零样本医疗疾病识别中的不足。
- LLaVA-RadZ采用解码侧特征对齐训练(DFAT),利用MLLM解码器架构的特点,并融入针对不同模态的模态特定令牌,实现图像和文本表征的有效利用和稳健的跨模态对齐。
- 引入领域知识锚定模块(DKAM),挖掘大型模型的内在医学知识,缩小图像-文本对齐中的类别语义差距。
- DKAM改进了类别级别的对齐,使得疾病识别更加准确。
- LLaVA-RadZ在多个基准测试中表现优异,显著优于传统MLLMs。
点此查看论文截图

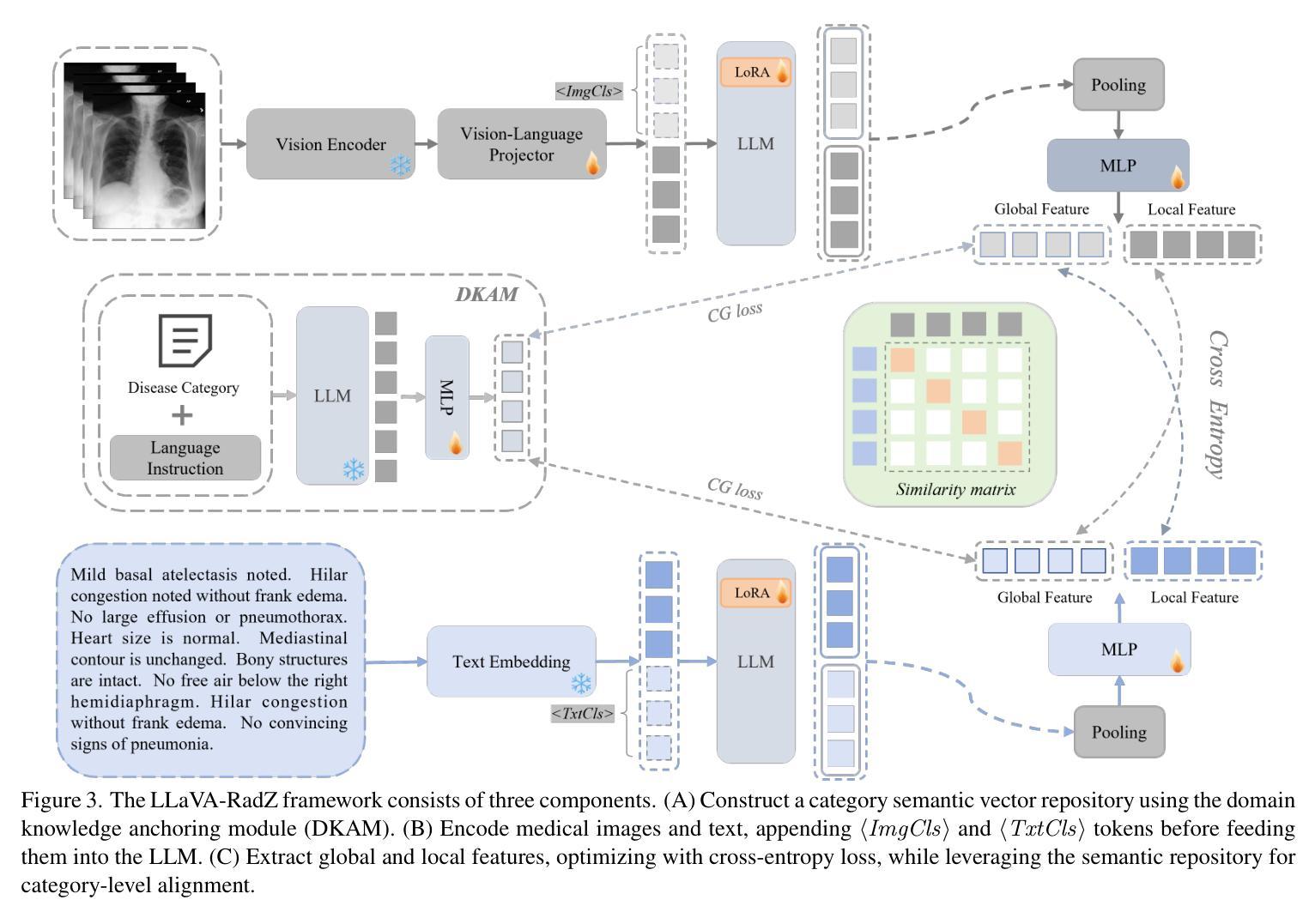
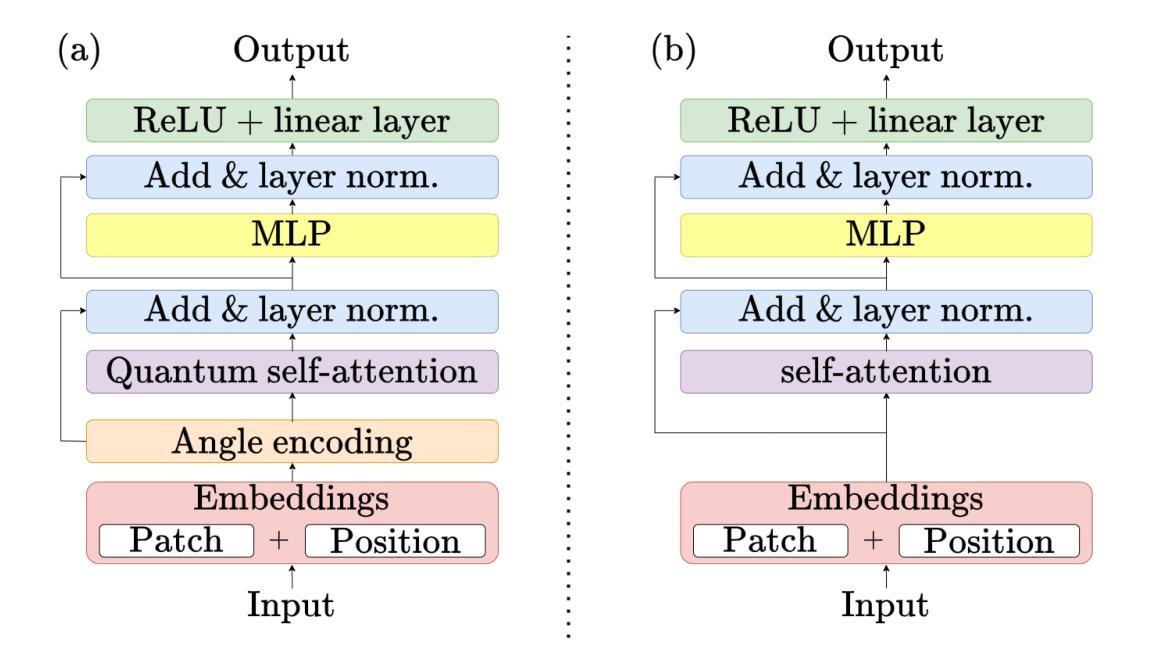
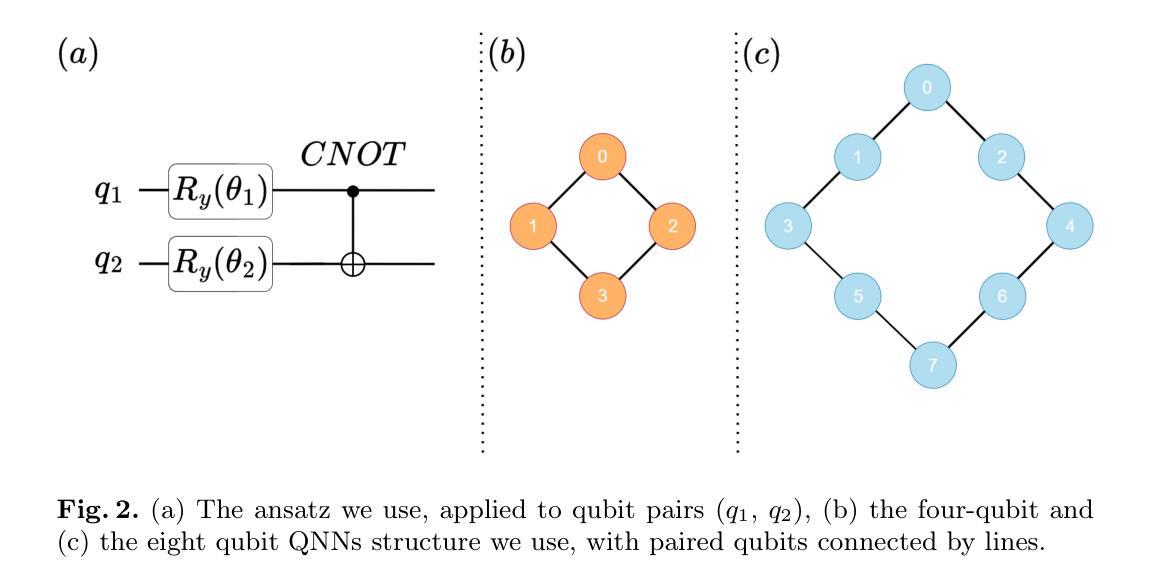
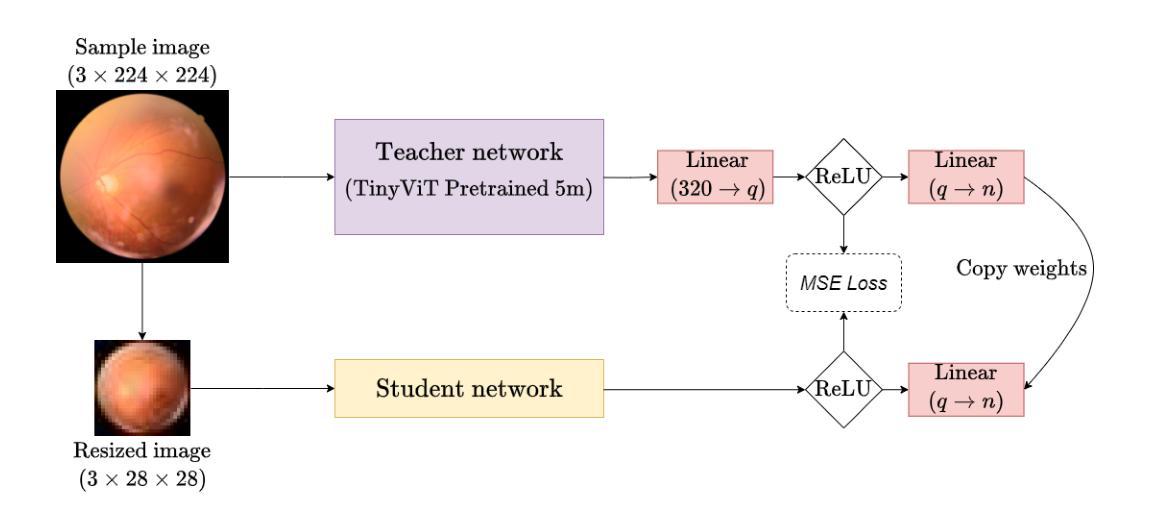
Distilling Knowledge into Quantum Vision Transformers for Biomedical Image Classification
Authors:Thomas Boucher, Evangelos B. Mazomenos
Quantum vision transformers (QViTs) build on vision transformers (ViTs) by replacing linear layers within the self-attention mechanism with parameterised quantum neural networks (QNNs), harnessing quantum mechanical properties to improve feature representation. This hybrid approach aims to achieve superior performance, with significantly reduced model complexity as a result of the enriched feature representation, requiring fewer parameters. This paper proposes a novel QViT model for biomedical image classification and investigates its performance against comparable ViTs across eight diverse datasets, encompassing various modalities and classification tasks. We assess models trained from scratch and those pre-trained using knowledge distillation (KD) from high-quality teacher models. Our findings demonstrate that QViTs outperform comparable ViTs with average ROC AUC (0.863 vs 0.846) and accuracy (0.710 vs 0.687) when trained from scratch, and even compete with state-of-the-art classical models in multiple tasks, whilst being significantly more efficient (89% reduction in GFLOPs and 99.99% in parameter number). Additionally, we find that QViTs and ViTs respond equally well to KD, with QViT pre-training performance scaling with model complexity. This is the first investigation into the efficacy of deploying QViTs with KD for computer-aided diagnosis. Our results highlight the enormous potential of quantum machine learning (QML) in biomedical image analysis.
量子视觉转换器(QViT)通过构建在视觉转换器(ViT)的基础上,将自注意力机制中的线性层替换为参数化量子神经网络(QNN),利用量子机械特性改进特征表示。这种混合方法旨在实现卓越的性能,并因特征表示更丰富而大大减少模型复杂度,从而减少了参数需求。本文提出了一种新型的QViT模型,用于生物医学图像分类,并研究了它在八个不同数据集上与相应的ViT的性能对比。这八个数据集涵盖多种模态和分类任务。我们评估了从头开始训练的模型以及使用来自高质量教师模型的蒸馏(KD)进行预训练的模型。我们的研究结果表明,从头开始训练的QViT在平均ROC AUC(0.863 vs 0.846)和准确度(0.710 vs 0.687)上超过了相应的ViT,甚至在多个任务中与最先进的经典模型竞争,同时效率更高(GFLOPs减少89%,参数数量减少99.99%)。此外,我们发现QViT和ViT对KD的响应同样良好,QViT的预训练性能随模型复杂度的增加而提高。这是首次调查将QViT与KD结合用于计算机辅助诊断的有效性。我们的结果突显了量子机器学习(QML)在生物医学图像分析中的巨大潜力。
论文及项目相关链接
PDF Submitted for MICCAI 2025
Summary
基于量子计算机算法理论的量子视觉变压器(QViT)技术取得了重大突破。此技术基于现有视觉变压器(ViT),并利用量子神经网络强化其特征表征功能。最新研究发现这种新技术可在显著减少模型复杂性的同时提高性能,尤其适用于生物医学图像分类任务。在多种数据集上进行的测试表明,使用QViT模型的分类器表现优于传统模型,尤其在高效计算方面有巨大优势。同时,本文也是首次探讨将QViT技术与知识蒸馏技术相结合应用于计算机辅助诊断的可能性。研究结果为量子机器学习在生物医学图像分析领域的未来应用开辟了新的方向。
Key Takeaways
- 量子视觉变压器(QViT)结合了量子神经网络与视觉变压器技术。它通过优化ViT的自注意力机制来增强其性能,从而提高模型的表征能力。
- QViT显著减少了模型的复杂性,并在多项生物医学图像分类任务中表现出卓越性能。在多个数据集上的实验结果表明,使用QViT的分类器性能优于传统模型。尤其是与传统的ViT相比,其平均ROC AUC和准确率均有显著提升。同时其能效极高,与传统模型相比大幅降低计算复杂度。
- QViT模型对知识蒸馏技术(KD)的响应良好,预训练性能随模型复杂度的增加而提升。这为未来计算机诊断中的模型训练提供了新的思路。这是首次尝试将QViT与KD结合应用于计算机辅助诊断的研究。结合两者能够提高模型训练的效率和效果。通过本次试验发现揭示了量子机器学习在生物医学图像分析领域巨大的应用潜力,证明融合不同技术和领域的合作前景十分光明。我们有望通过高效算法实现更准确的诊断。
点此查看论文截图

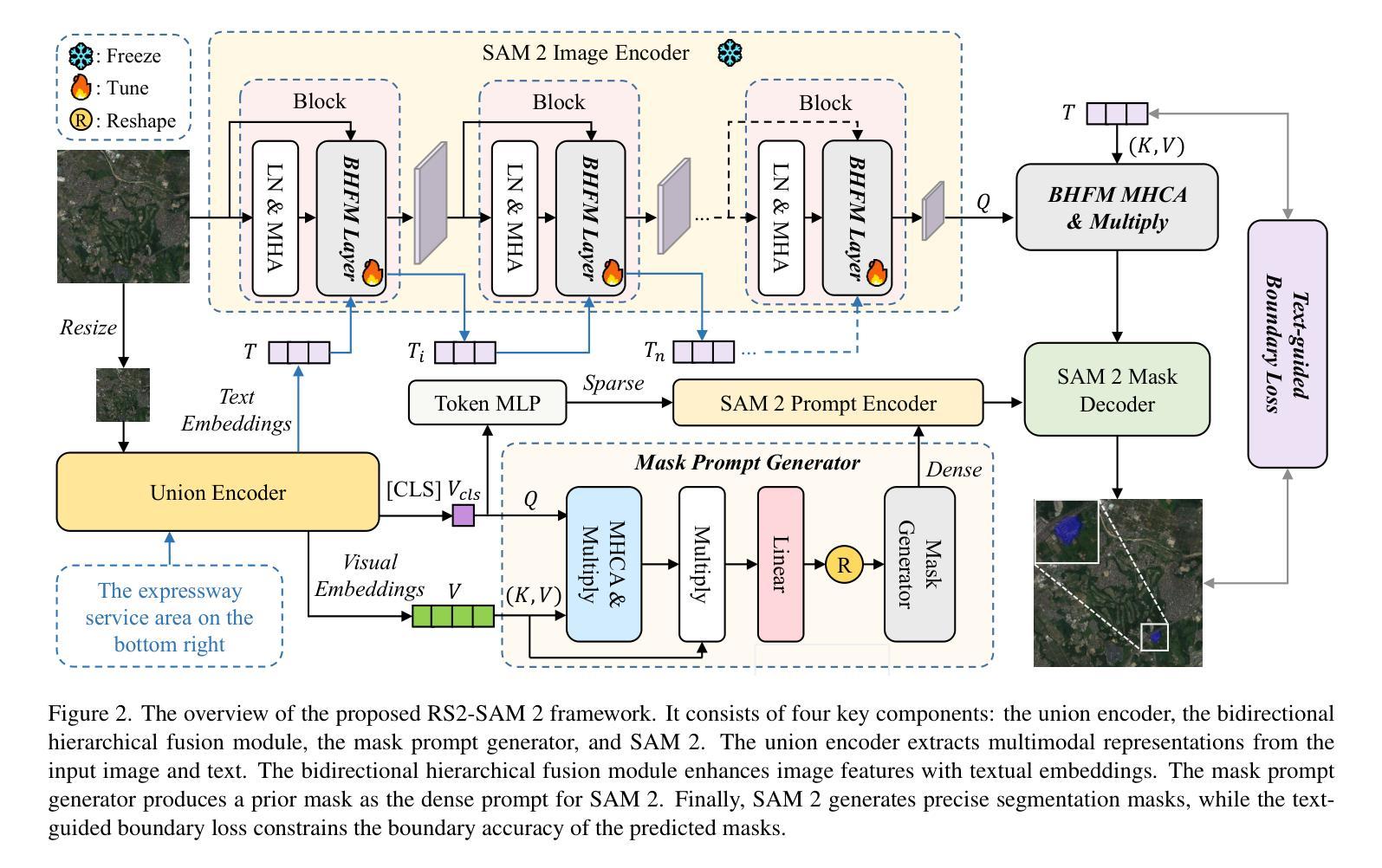
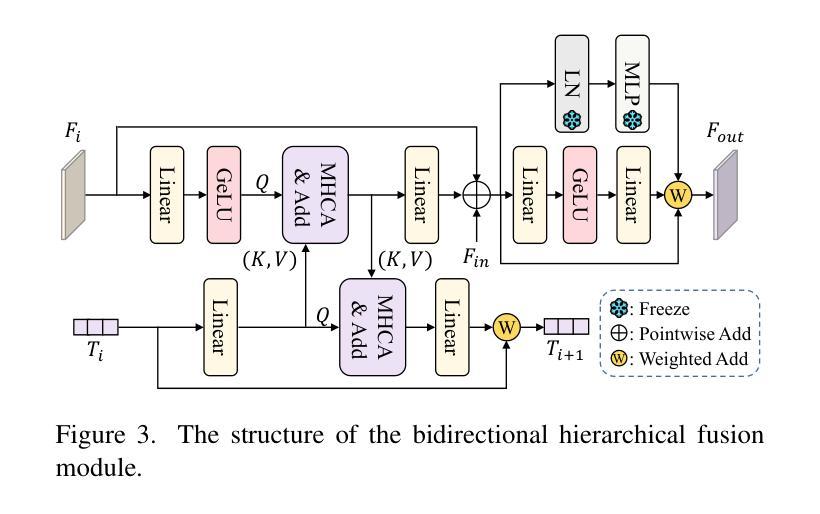
Customized SAM 2 for Referring Remote Sensing Image Segmentation
Authors:Fu Rong, Meng Lan, Qian Zhang, Lefei Zhang
Referring Remote Sensing Image Segmentation (RRSIS) aims to segment target objects in remote sensing (RS) images based on textual descriptions. Although Segment Anything Model 2 (SAM 2) has shown remarkable performance in various segmentation tasks, its application to RRSIS presents several challenges, including understanding the text-described RS scenes and generating effective prompts from text descriptions. To address these issues, we propose RS2-SAM 2, a novel framework that adapts SAM 2 to RRSIS by aligning the adapted RS features and textual features, providing pseudo-mask-based dense prompts, and enforcing boundary constraints. Specifically, we first employ a union encoder to jointly encode the visual and textual inputs, generating aligned visual and text embeddings as well as multimodal class tokens. Then, we design a bidirectional hierarchical fusion module to adapt SAM 2 to RS scenes and align adapted visual features with the visually enhanced text embeddings, improving the model’s interpretation of text-described RS scenes. Additionally, a mask prompt generator is introduced to take the visual embeddings and class tokens as input and produce a pseudo-mask as the dense prompt of SAM 2. To further refine segmentation, we introduce a text-guided boundary loss to optimize segmentation boundaries by computing text-weighted gradient differences. Experimental results on several RRSIS benchmarks demonstrate that RS2-SAM 2 achieves state-of-the-art performance.
远程遥感图像分割(RRSIS)旨在根据文本描述对遥感(RS)图像中的目标对象进行分割。尽管Segment Anything Model 2(SAM 2)在各种分割任务中表现出卓越的性能,但将其应用于RRSIS面临一些挑战,包括理解文本描述的RS场景和从文本描述中产生有效的提示。为了解决这些问题,我们提出了RS2-SAM 2,这是一个新型框架,通过适应SAM 2到RRSIS,通过对齐适应的RS特征和文本特征、提供基于伪掩码的密集提示和执行边界约束来适应SAM 2。具体来说,我们首先采用联合编码器对视觉和文本输入进行编码,生成对齐的视觉和文本嵌入以及多模态类标记。然后,我们设计了一个双向层次融合模块,使SAM 2适应RS场景,并使适应的视觉特征与增强的文本嵌入对齐,提高模型对文本描述的RS场景的解释能力。此外,引入了掩膜提示生成器,以视觉嵌入和类标记为输入,生成伪掩膜作为SAM 2的密集提示。为了进一步细化分割,我们引入了一种文本引导边界损失,通过计算文本加权梯度差异来优化分割边界。在多个RRSIS基准测试上的实验结果表明,RS2-SAM 2达到了最新技术水平。
论文及项目相关链接
Summary
远程感应图像分割的文本描述(RRSIS)旨在根据文本描述对遥感图像的目标对象进行分割。针对SAM 2模型在RRSIS应用中的挑战,我们提出了RS2-SAM 2框架,通过结合遥感特征和文本特征、提供基于伪掩码的密集提示并强制执行边界约束,以适应RRSIS。实验结果表明,RS2-SAM 2在多个RRSIS基准测试中取得了最佳性能。
Key Takeaways
- RS2-SAM 2旨在解决SAM 2在遥感图像分段中的文本描述应用中的挑战。
- 通过联合编码视觉和文本输入,生成对齐的视觉和文本嵌入以及多模态类令牌,以理解文本描述的遥感场景。
- 设计的双向层次融合模块使SAM 2适应遥感场景,将适应的视觉特征与视觉增强的文本嵌入对齐。
- 引入掩膜提示生成器,以视觉嵌入和类令牌为输入,生成伪掩码作为SAM 2的密集提示。
- 通过计算文本加权的梯度差异来优化分割边界,进一步细化分割。
- RS2-SAM 2在多个遥感图像分割基准测试中实现了最佳性能。
点此查看论文截图

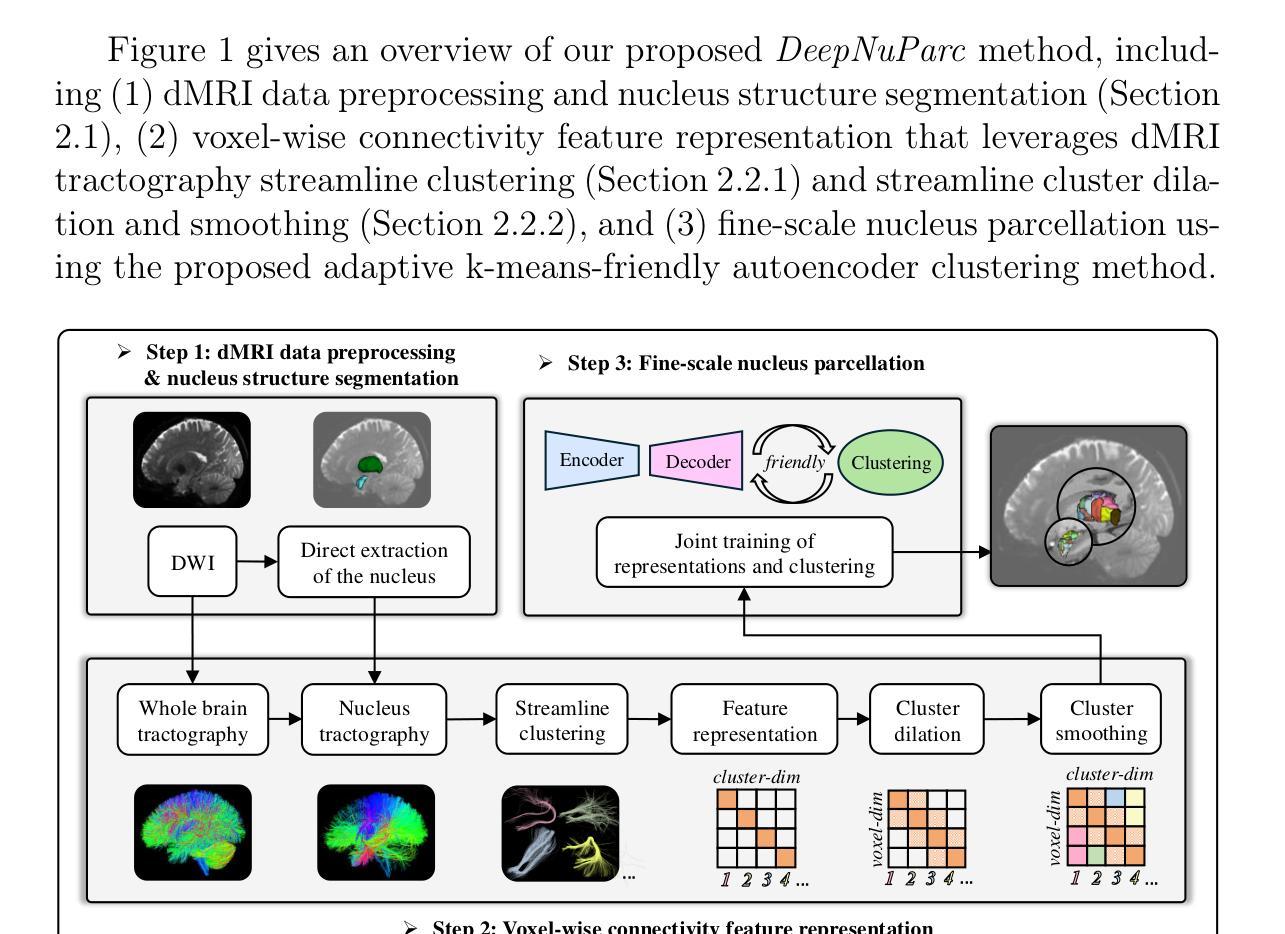
DeepNuParc: A Novel Deep Clustering Framework for Fine-scale Parcellation of Brain Nuclei Using Diffusion MRI Tractography
Authors:Haolin He, Ce Zhu, Le Zhang, Yipeng Liu, Xiao Xu, Yuqian Chen, Leo Zekelman, Jarrett Rushmore, Yogesh Rathi, Nikos Makris, Lauren J. O’Donnell, Fan Zhang
Brain nuclei are clusters of anatomically distinct neurons that serve as important hubs for processing and relaying information in various neural circuits. Fine-scale parcellation of the brain nuclei is vital for a comprehensive understanding of its anatomico-functional correlations. Diffusion MRI tractography is an advanced imaging technique that can estimate the brain’s white matter structural connectivity to potentially reveal the topography of the nuclei of interest for studying its subdivisions. In this work, we present a deep clustering pipeline, namely DeepNuParc, to perform automated, fine-scale parcellation of brain nuclei using diffusion MRI tractography. First, we incorporate a newly proposed deep learning approach to enable accurate segmentation of the nuclei of interest directly on the dMRI data. Next, we design a novel streamline clustering-based structural connectivity feature for a robust representation of voxels within the nuclei. Finally, we improve the popular joint dimensionality reduction and k-means clustering approach to enable nuclei parcellation at a finer scale. We demonstrate DeepNuParc on two important brain structures, i.e. the amygdala and the thalamus, that are known to have multiple anatomically and functionally distinct nuclei subdivisions. Experimental results show that DeepNuParc enables consistent parcellation of the nuclei into multiple parcels across multiple subjects and achieves good correspondence with the widely used coarse-scale atlases. Our codes are available at https://github.com/HarlandZZC/deep_nuclei_parcellation.
脑核是由解剖结构独特的神经元聚集而成的重要枢纽,用于处理和传递各种神经网络中的信息。对脑核进行精细的分区对于全面理解其解剖功能关联至关重要。扩散MRI追踪成像是一种先进的成像技术,可以估计大脑的白色物质结构连接性,以揭示感兴趣核团的拓扑结构,从而研究其亚区。在这项工作中,我们提出了一种深度聚类流程,即DeepNuParc,使用扩散MRI追踪成像技术自动执行脑核的精细分区。首先,我们采用新提出的深度学习方法进行准确分割,直接在dMRI数据上对感兴趣的核团进行分割。接下来,我们设计了一种基于流线聚类的结构连接特征,以稳健地表示核团内的体素。最后,我们改进了流行的联合降维和K均值聚类方法,以实现更精细的核团分区。我们在两个重要的脑结构——杏仁核和丘脑上展示了DeepNuParc的应用,这两个结构已知具有多个解剖和功能上独特的核团亚区。实验结果表明,DeepNuParc能够在多个受试者中对核团进行一致的精细分割,并与广泛使用的粗略图谱实现了良好的对应。我们的代码可从 https://github.com/HarlandZZC/deep_nuclei_parcellation 获取。
论文及项目相关链接
Summary
本文介绍了一种名为DeepNuParc的深度学习驱动的大脑核团精细分割方法,该方法结合了扩散MRI成像技术和深度学习方法,实现对大脑核团进行自动化精细分割。DeepNuParc可应用于多个已知具有多个解剖和功能上独特核团的大脑结构,如杏仁核和丘脑。该方法在多个受试者之间实现了稳定的核团分割,并与广泛使用的粗尺度图谱具有良好的对应关系。
Key Takeaways
- 文中提到了Brain nuclei作为处理信息和传递信息的重要枢纽,其精细分割对理解解剖功能关联至关重要。
- 采用扩散MRI成像技术估计大脑白质结构连接性,以揭示核团拓扑结构。
- 提出了一种深度学习驱动的大脑核团精细分割方法——DeepNuParc。
- DeepNuParc结合了深度学习和扩散MRI数据,实现了对大脑核团的直接分割。
- 通过流线聚类技术,DeepNuParc可有效地表示核团内的体素特征。
- 实验结果表明,DeepNuParc可实现跨多个受试者的核团一致性分割,并与粗尺度图谱有良好的对应关系。
点此查看论文截图

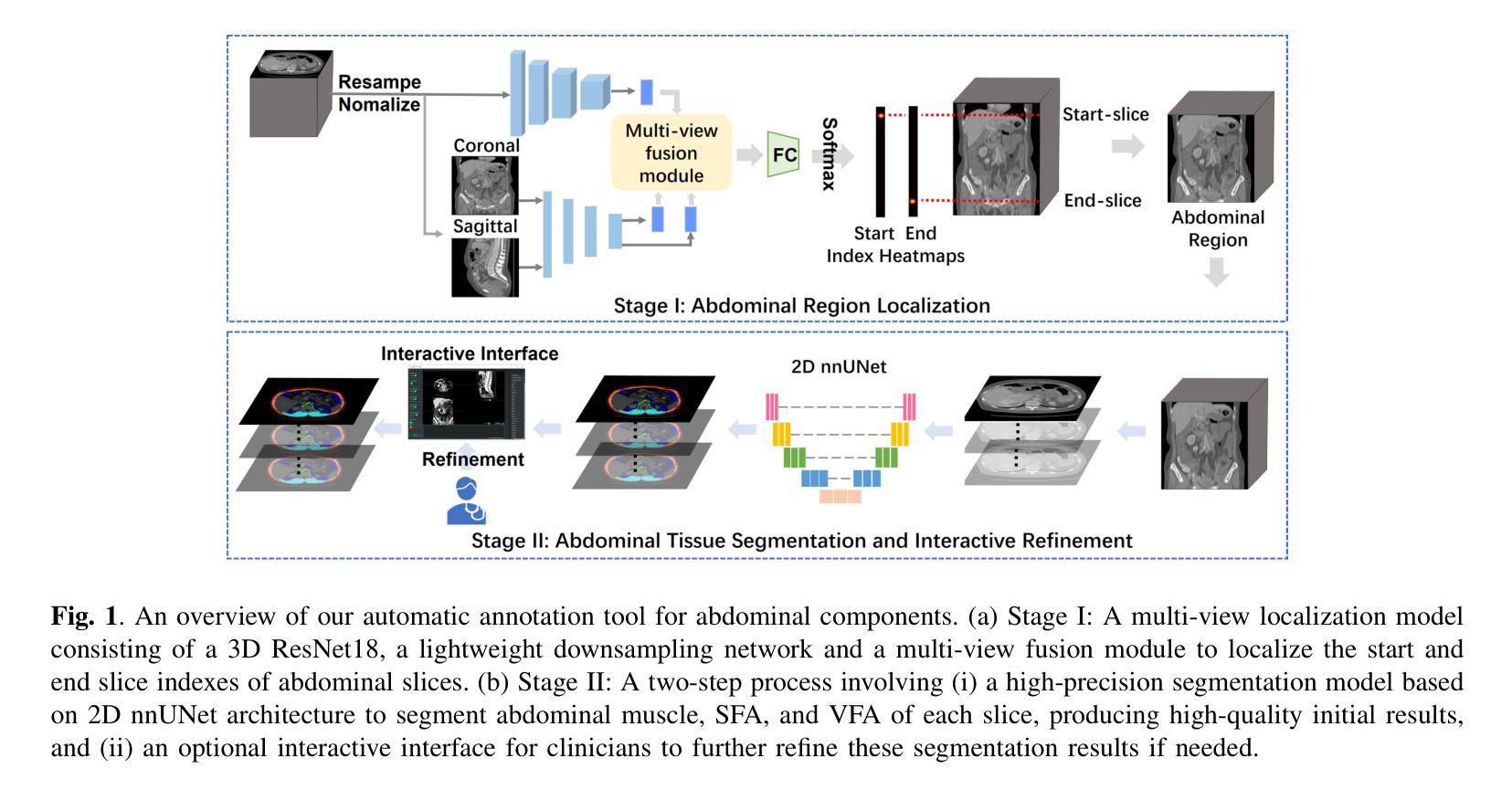
AI-Driven Automated Tool for Abdominal CT Body Composition Analysis in Gastrointestinal Cancer Management
Authors:Xinyu Nan, Meng He, Zifan Chen, Bin Dong, Lei Tang, Li Zhang
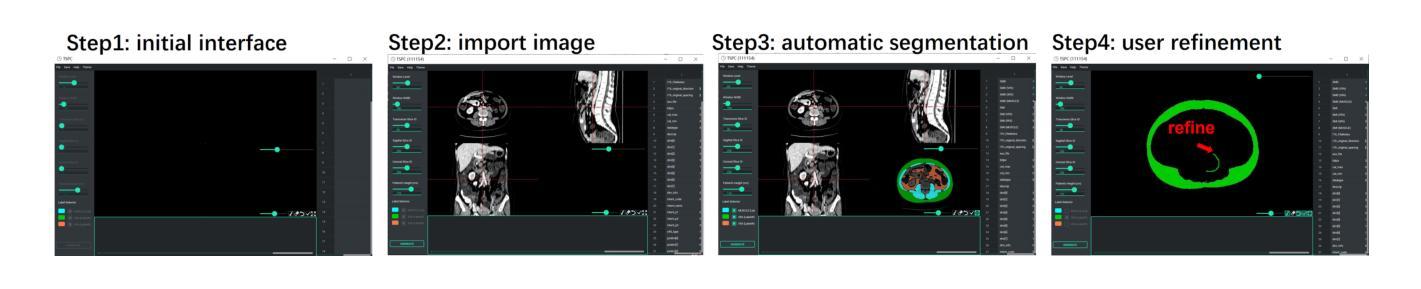
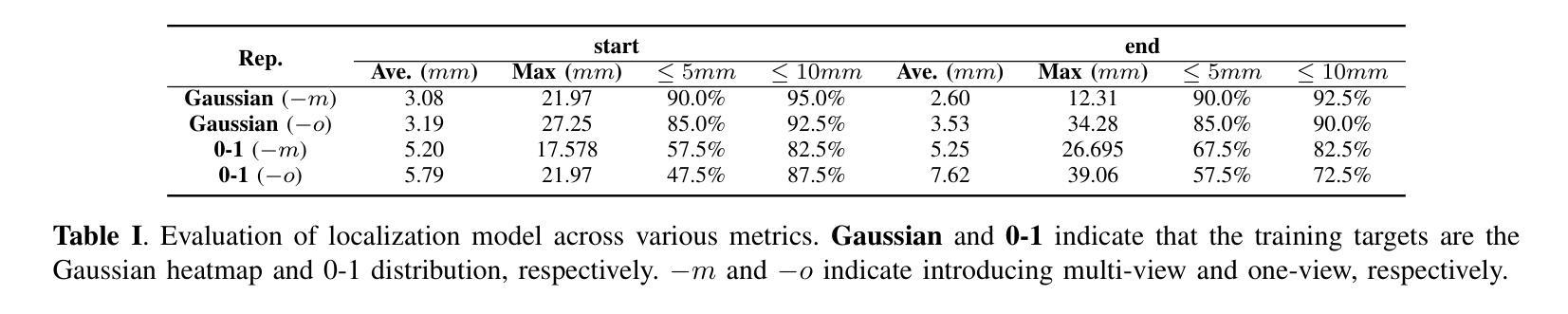
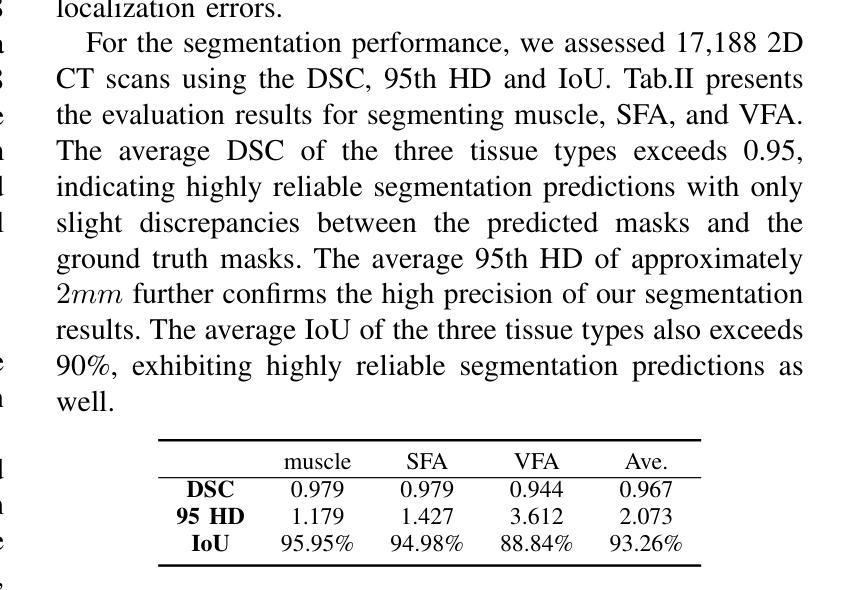
The incidence of gastrointestinal cancers remains significantly high, particularly in China, emphasizing the importance of accurate prognostic assessments and effective treatment strategies. Research shows a strong correlation between abdominal muscle and fat tissue composition and patient outcomes. However, existing manual methods for analyzing abdominal tissue composition are time-consuming and costly, limiting clinical research scalability. To address these challenges, we developed an AI-driven tool for automated analysis of abdominal CT scans to effectively identify and segment muscle, subcutaneous fat, and visceral fat. Our tool integrates a multi-view localization model and a high-precision 2D nnUNet-based segmentation model, demonstrating a localization accuracy of 90% and a Dice Score Coefficient of 0.967 for segmentation. Furthermore, it features an interactive interface that allows clinicians to refine the segmentation results, ensuring high-quality outcomes effectively. Our tool offers a standardized method for effectively extracting critical abdominal tissues, potentially enhancing the management and treatment for gastrointestinal cancers. The code is available at https://github.com/NanXinyu/AI-Tool4Abdominal-Seg.git}{https://github.com/NanXinyu/AI-Tool4Abdominal-Seg.git.
胃肠道癌症的发病率仍然很高,特别是在中国,这强调了准确预后评估和有效治疗策略的重要性。研究表明,腹部肌肉和脂肪组织成分与患者的治疗效果之间存在强烈的相关性。然而,现有的分析腹部组织成分的手工方法既耗时又成本高昂,限制了临床研究的可扩展性。为了应对这些挑战,我们开发了一种人工智能驱动的工具,用于自动分析腹部CT扫描结果,有效识别并分割肌肉、皮下脂肪和内脏脂肪。我们的工具集成了一个多视图定位模型和一个基于高精度二维nnUNet的分割模型,定位精度达到90%,分割的Dice系数达到0.967。此外,它还具有一个交互式界面,允许临床医生对分割结果进行微调,确保高质量的结果。我们的工具提供了一种有效提取关键腹部组织的方法,可能有助于胃肠道癌症的管理和治疗。代码可在https://github.com/NanXinyu/AI-Tool4Abdominal-Seg.git上获取。
论文及项目相关链接
Summary
本文主要介绍了中国胃肠道癌症的发病率仍然较高,强调了对患者进行准确的预后评估和有效治疗策略的重要性。为解决现有分析腹部组织成分方法耗时且成本高昂的问题,研究团队开发了一种基于人工智能的工具,可自动分析腹部CT扫描图像,有效识别并分割肌肉、皮下脂肪和内脏脂肪。该工具结合了多视角定位模型和基于高精度二维nnUNet的分割模型,定位准确率高达90%,分割的Dice系数达到0.967。此外,其交互界面使临床医生能够优化分割结果,确保高质量的结果输出。此工具为标准化提取关键腹部组织提供了有效方法,有望改善胃肠道癌症的管理和治疗。
Key Takeaways
- 胃肠道癌症在中国发病率高,需要准确的预后评估和有效治疗策略。
- 现有分析腹部组织成分的方法耗时且成本高昂,限制了临床研究的发展。
- 研究团队开发了一种基于AI的工具,可以自动分析腹部CT扫描图像,识别并分割肌肉、皮下脂肪和内脏脂肪。
- 该工具采用多视角定位模型和基于高精度二维nnUNet的分割模型,具有高的定位准确率和分割性能。
- 工具的交互界面允许临床医生优化分割结果,确保高质量的结果输出。
- 此工具为标准化提取关键腹部组织提供了有效方法。
点此查看论文截图

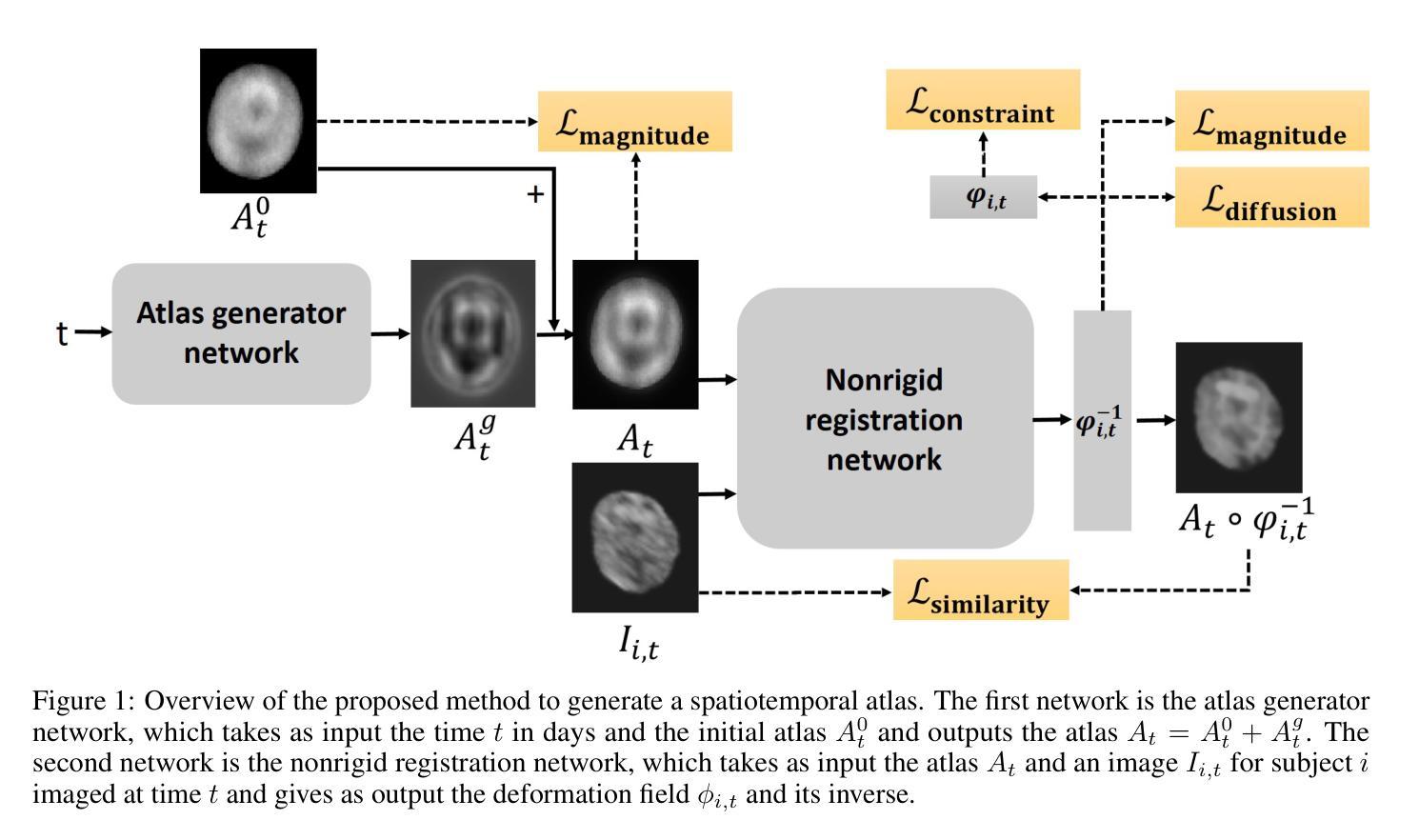
The 4D Human Embryonic Brain Atlas: spatiotemporal atlas generation for rapid anatomical changes using first-trimester ultrasound from the Rotterdam Periconceptional Cohort
Authors:Wietske A. P. Bastiaansen, Melek Rousian, Anton H. J. Koning, Wiro J. Niessen, Bernadette S. de Bakker, Régine P. M. Steegers-Theunissen, Stefan Klein
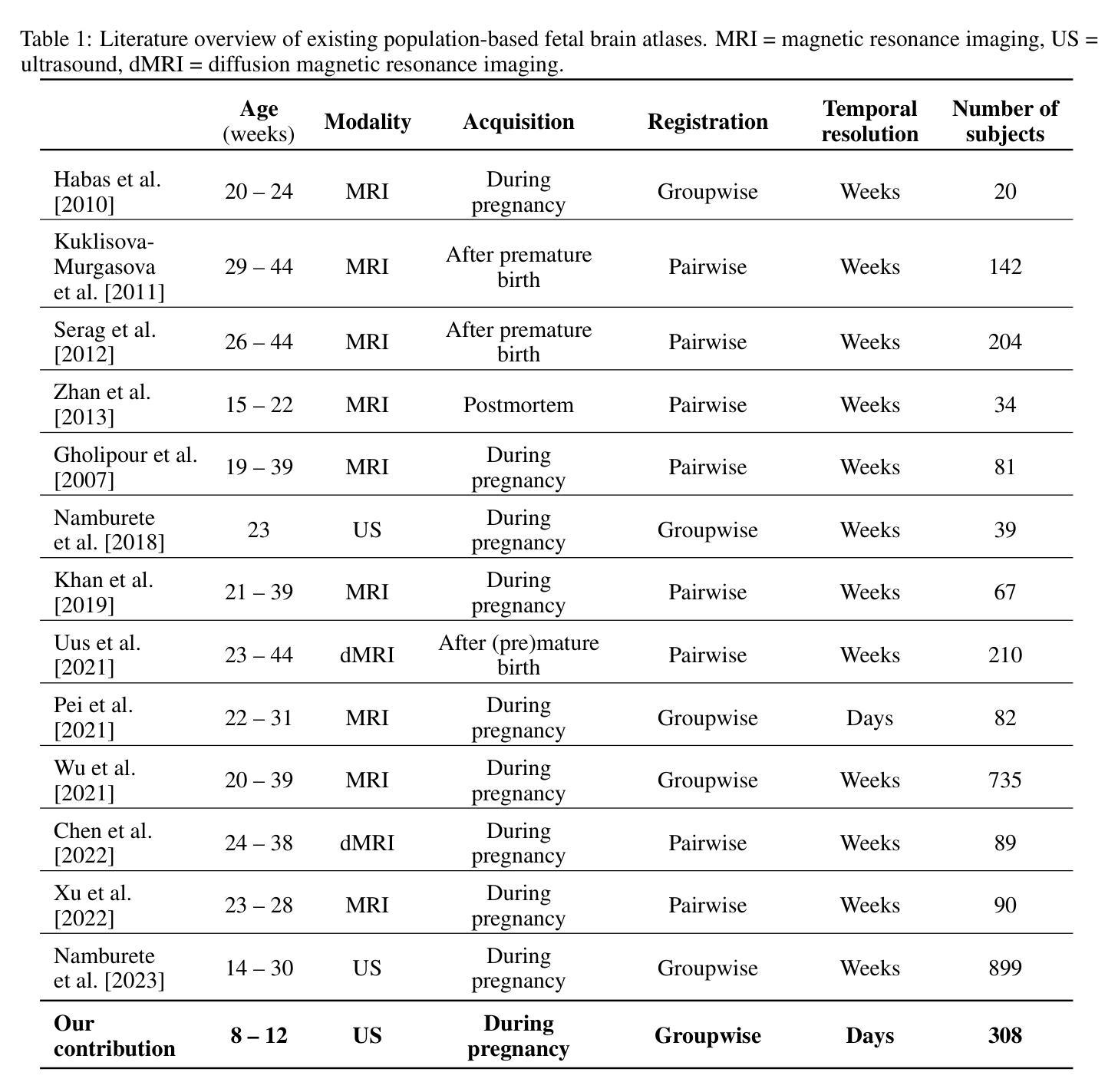
Early brain development is crucial for lifelong neurodevelopmental health. However, current clinical practice offers limited knowledge of normal embryonic brain anatomy on ultrasound, despite the brain undergoing rapid changes within the time-span of days. To provide detailed insights into normal brain development and identify deviations, we created the 4D Human Embryonic Brain Atlas using a deep learning-based approach for groupwise registration and spatiotemporal atlas generation. Our method introduced a time-dependent initial atlas and penalized deviations from it, ensuring age-specific anatomy was maintained throughout rapid development. The atlas was generated and validated using 831 3D ultrasound images from 402 subjects in the Rotterdam Periconceptional Cohort, acquired between gestational weeks 8 and 12. We evaluated the effectiveness of our approach with an ablation study, which demonstrated that incorporating a time-dependent initial atlas and penalization produced anatomically accurate results. In contrast, omitting these adaptations led to anatomically incorrect atlas. Visual comparisons with an existing ex-vivo embryo atlas further confirmed the anatomical accuracy of our atlas. In conclusion, the proposed method successfully captures the rapid anotomical development of the embryonic brain. The resulting 4D Human Embryonic Brain Atlas provides a unique insights into this crucial early life period and holds the potential for improving the detection, prevention, and treatment of prenatal neurodevelopmental disorders.
早期大脑发育对终身神经发育健康至关重要。然而,尽管大脑在几天内经历快速变化,但目前的临床实践对超声下的正常胚胎大脑解剖结构了解有限。为了深入了解正常大脑发育并识别偏差,我们采用了基于深度学习的方法创建四维人类胚胎大脑图谱进行群体注册和时空图谱生成。我们的方法引入了一个时间依赖的初始图谱,并对偏离该图谱的情况进行惩罚,以确保在快速发育过程中保持特定年龄的解剖学特征。该图谱是使用来自鹿特丹胚胎期队列的402名受试者的831张三维超声图像生成和验证的,这些图像是在妊娠第8至第12周期间获取的。我们通过一项消融研究评估了我们的方法的有效性,该研究证明采用时间依赖的初始图谱和惩罚机制能够产生解剖结构准确的结果。相比之下,省略这些适应性调整会导致解剖结构不正确的图谱。与现有的体外胚胎图谱的视觉比较进一步证实了我们图谱的解剖学准确性。总之,所提出的方法成功捕捉了胚胎大脑的快速解剖学发育过程。所得的四维人类胚胎大脑图谱为这一关键早期生命阶段提供了独特的见解,并具有改善产前神经发育障碍检测、预防和治疗的潜力。
论文及项目相关链接
Summary
该研究使用深度学习技术对早期胚胎脑进行四维动态图像生成与分析,成功构建了首个四人类胚胎脑图谱。研究解决了超声扫描技术对胚胎脑解剖结构的准确解析问题,通过时间依赖的初始图谱与偏差惩罚机制确保了胚胎脑发育的快速变化过程中保持特定的年龄结构特征。该研究有助于深入理解胚胎脑发育过程,对产前神经发育障碍的预测和治疗具有潜在价值。
Key Takeaways
- 研究强调了早期大脑发育对终身神经发育健康的重要性,并指出了当前临床实践中对胚胎大脑发育知识了解的局限性。
- 研究使用深度学习技术创建了首个四维人类胚胎脑图谱,以深入了解正常的大脑发育情况并识别偏差。
- 该研究解决了超声扫描技术对胚胎脑解剖结构准确解析的问题,确保在胚胎脑快速发育过程中保持特定的年龄结构特征。
- 通过使用831张三维超声图像进行验证,证明了该方法的准确性和有效性。
- 研究通过消融实验证明,包含时间依赖的初始图谱和偏差惩罚机制的方法能生成解剖结构准确的图谱。
- 与现有的体外胚胎图谱的视觉比较进一步证实了该图谱的解剖学准确性。
点此查看论文截图

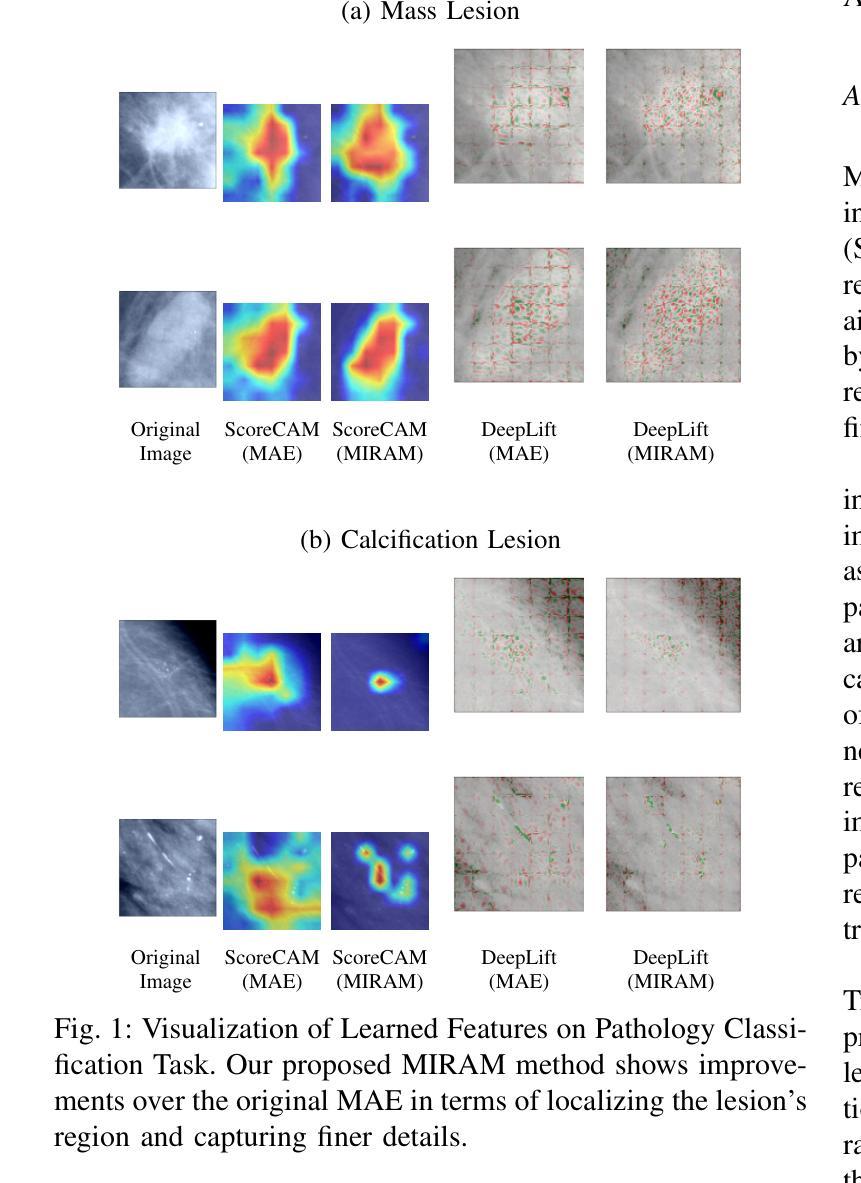
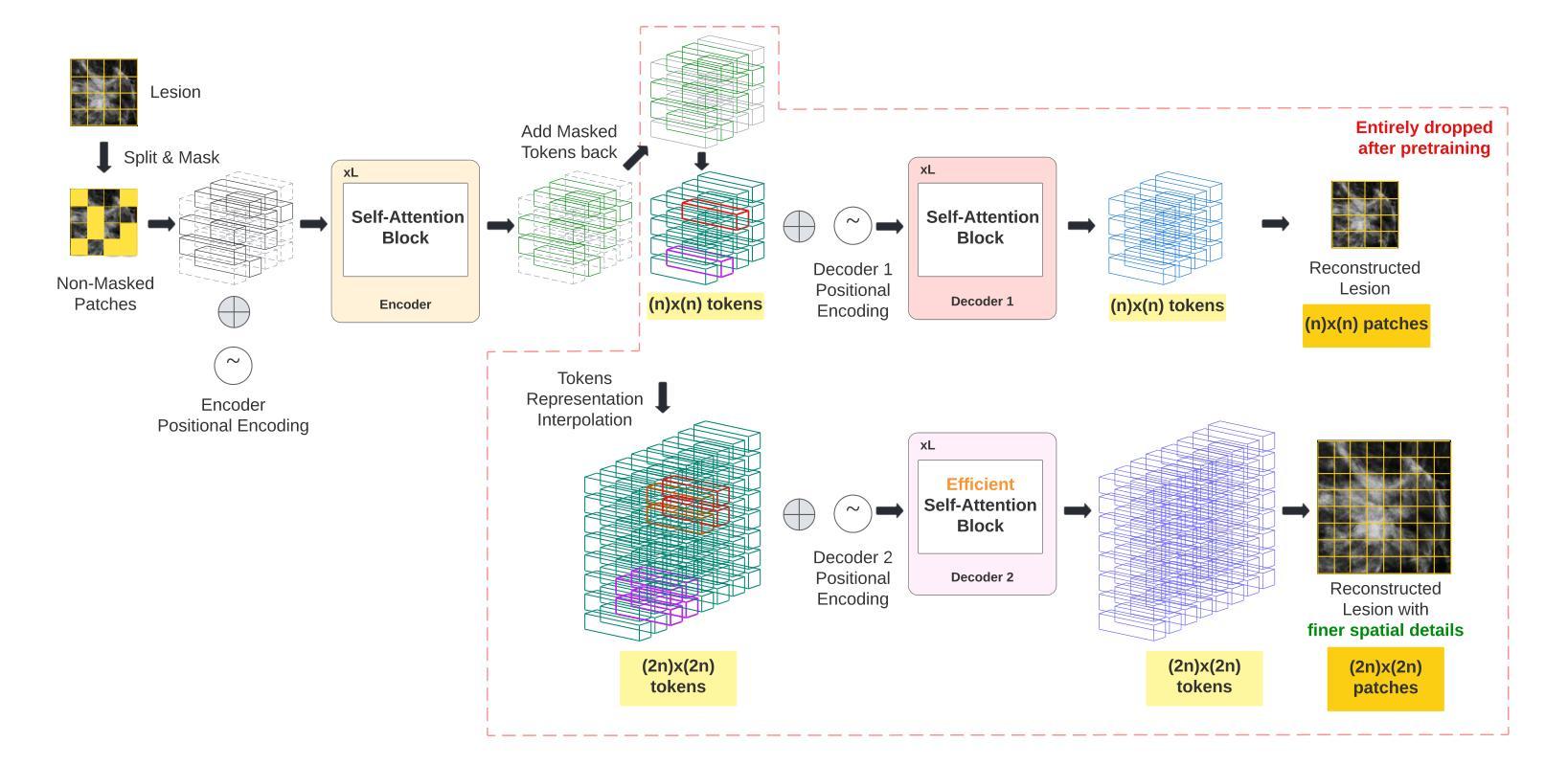
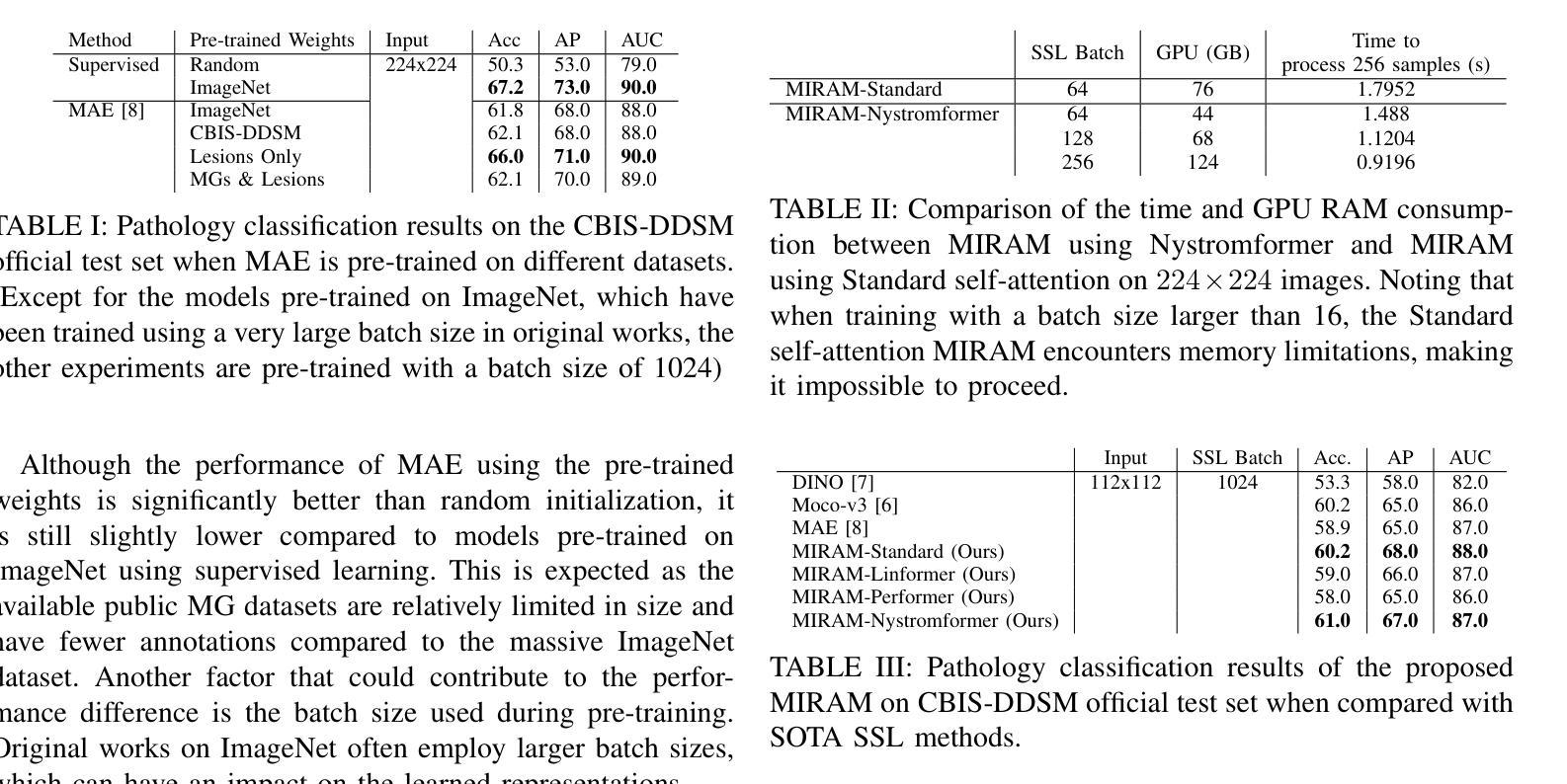
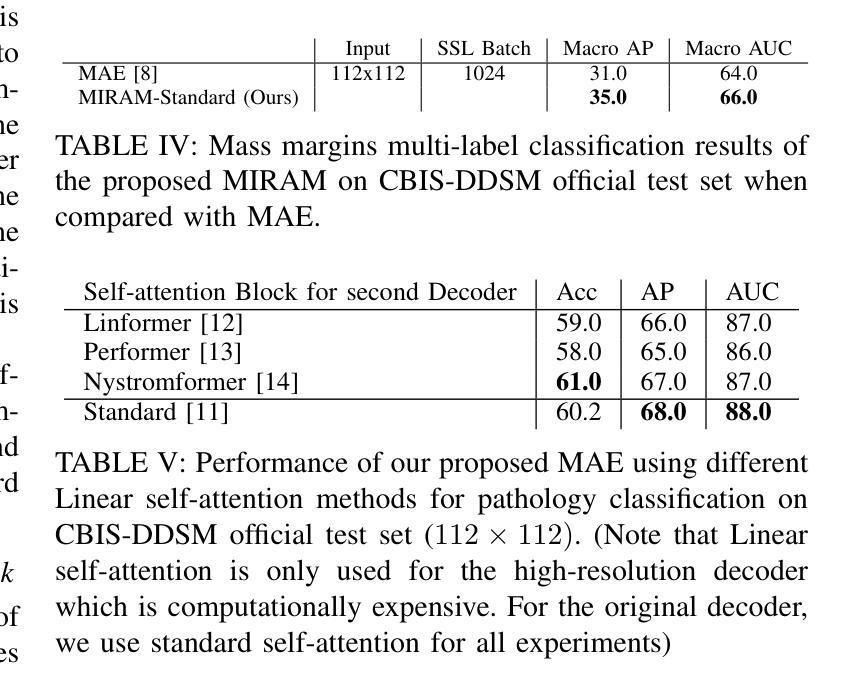
MIRAM: Masked Image Reconstruction Across Multiple Scales for Breast Lesion Risk Prediction
Authors:Hung Q. Vo, Pengyu Yuan, Zheng Yin, Kelvin K. Wong, Chika F. Ezeana, Son T. Ly, Stephen T. C. Wong, Hien V. Nguyen
Self-supervised learning (SSL) has garnered substantial interest within the machine learning and computer vision communities. Two prominent approaches in SSL include contrastive-based learning and self-distillation utilizing cropping augmentation. Lately, masked image modeling (MIM) has emerged as a more potent SSL technique, employing image inpainting as a pretext task. MIM creates a strong inductive bias toward meaningful spatial and semantic understanding. This has opened up new opportunities for SSL to contribute not only to classification tasks but also to more complex applications like object detection and image segmentation. Building upon this progress, our research paper introduces a scalable and practical SSL approach centered around more challenging pretext tasks that facilitate the acquisition of robust features. Specifically, we leverage multi-scale image reconstruction from randomly masked input images as the foundation for feature learning. Our hypothesis posits that reconstructing high-resolution images enables the model to attend to finer spatial details, particularly beneficial for discerning subtle intricacies within medical images. The proposed SSL features help improve classification performance on the Curated Breast Imaging Subset of Digital Database for Screening Mammography (CBIS-DDSM) dataset. In pathology classification, our method demonstrates a 3% increase in average precision (AP) and a 1% increase in the area under the receiver operating characteristic curve (AUC) when compared to state-of-the-art (SOTA) algorithms. Moreover, in mass margins classification, our approach achieves a 4% increase in AP and a 2% increase in AUC.
自监督学习(SSL)在机器学习和计算机视觉领域引起了极大的关注。自监督学习的两种主要方法包括基于对比学习和使用裁剪增强的自我蒸馏。最近,基于图像补全的遮罩图像建模(MIM)作为一种更强大的SSL技术而出现。MIM对有意义的空间和语义理解产生了强烈的归纳偏见。这为SSL不仅为分类任务做出贡献,也为更复杂的任务如目标检测和图像分割等提供了新机会。基于这一进展,我们的研究论文介绍了一种可扩展且实用的SSL方法,该方法以更具挑战性的预文本任务为中心,促进稳健特征的获取。具体来说,我们利用从随机遮罩的输入图像进行多尺度图像重建作为特征学习的基础。我们的假设是,重建高分辨率图像可以使模型关注更精细的空间细节,这对识别医学图像中的微妙细节特别有益。所提出的SSL特征有助于改进数字数据库筛选乳腺摄影术的精选乳腺成像子集(CBIS-DDSM)数据集上的分类性能。在病理分类方面,与最先进的算法相比,我们的方法在平均精确度上提高了3%,在接收特性曲线下面积(AUC)上提高了1%。此外,在肿块边缘分类中,我们的方法实现了平均精确度提高4%,AUC提高2%。
论文及项目相关链接
Summary
本文介绍了自监督学习(SSL)在机器学习和计算机视觉领域中的最新研究进展。重点介绍了一种新型的SSL方法,通过多尺度图像重建作为特征学习的基础,利用随机掩码输入图像进行重建。该方法在医学图像分类任务中表现出优异性能,特别是在乳腺图像分类、病理分类和肿瘤边缘分类等方面,相较于现有算法有所提升。
Key Takeaways
- 自监督学习(SSL)已成为机器学习和计算机视觉领域的热门研究方向。
- 新型SSL方法利用多尺度图像重建作为特征学习基础,通过随机掩码输入图像进行重建。
- 该方法能有效获取稳健特征,不仅适用于分类任务,还可应用于更复杂的应用,如目标检测和图像分割。
- 假设高分辨率图像重建能促使模型关注更精细的空间细节,对辨识医学图像中的微妙细节特别有益。
- 在乳腺图像分类方面,新方法相较于现有算法在平均精度(AP)上提高了3%,在受试者工作特征曲线下面积(AUC)上提高了1%。
- 在病理分类方面,新方法的AP提高了3%,AUC提高了1%。
点此查看论文截图

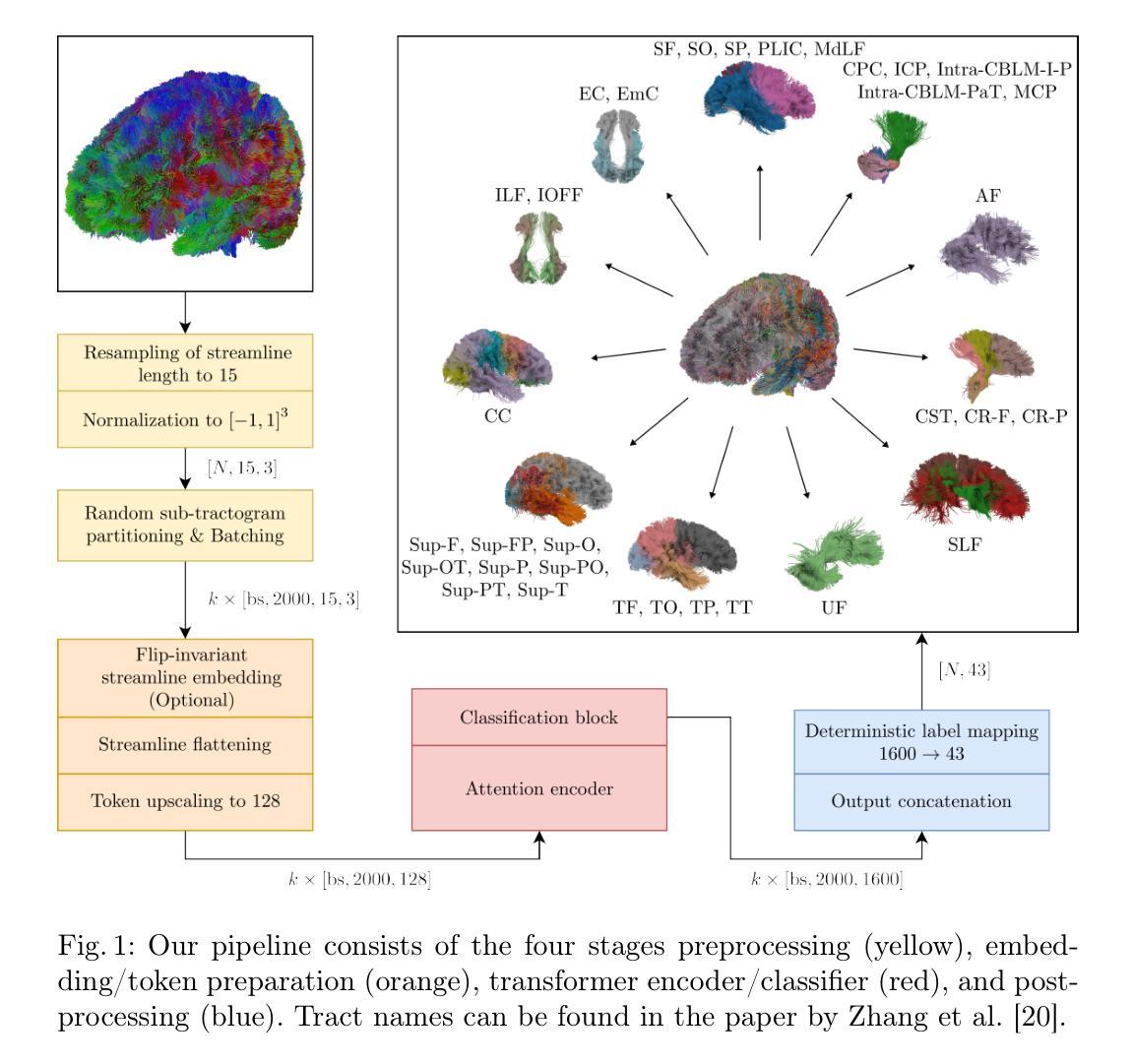
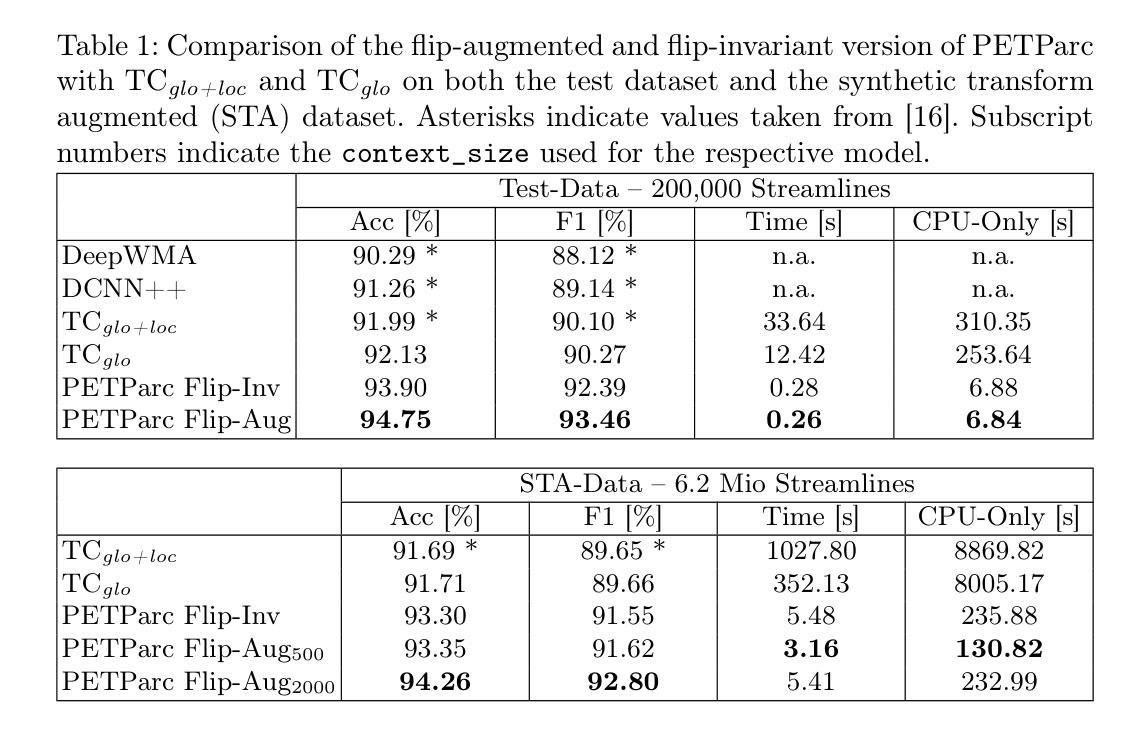
Global Context Is All You Need for Parallel Efficient Tractography Parcellation
Authors:Valentin von Bornhaupt, Johannes Grün, and Justus Bisten, Tobias Bauer, Theodor Rüber, Thomas Schultz
Whole-brain tractography in diffusion MRI is often followed by a parcellation in which each streamline is classified as belonging to a specific white matter bundle, or discarded as a false positive. Efficient parcellation is important both in large-scale studies, which have to process huge amounts of data, and in the clinic, where computational resources are often limited. TractCloud is a state-of-the-art approach that aims to maximize accuracy with a local-global representation. We demonstrate that the local context does not contribute to the accuracy of that approach, and is even detrimental when dealing with pathological cases. Based on this observation, we propose PETParc, a new method for Parallel Efficient Tractography Parcellation. PETParc is a transformer-based architecture in which the whole-brain tractogram is randomly partitioned into sub-tractograms whose streamlines are classified in parallel, while serving as global context for each other. This leads to a speedup of up to two orders of magnitude relative to TractCloud, and permits inference even on clinical workstations without a GPU. PETParc accounts for the lack of streamline orientation either via a novel flip-invariant embedding, or by simply using flips as part of data augmentation. Despite the speedup, results are often even better than those of prior methods. The code and pretrained model will be made public upon acceptance.
在扩散MRI的全脑轨迹成像后通常会进行分区,将每条流线分类为特定的白质束或剔除为假阳性。在大规模研究和临床中,高效分区都非常重要,因为必须处理大量数据且计算资源通常有限。TractCloud是一种前沿方法,旨在通过局部全局表示法来最大化准确性。我们证明,局部上下文并不有助于该方法的准确性,在处理病理病例时甚至是有害的。基于这一观察,我们提出了PETParc,这是一种新的并行高效轨迹分割方法。PETParc是基于transformer的架构,将全脑轨迹图随机分割成子轨迹图,流线分类并行进行,同时相互作为全局上下文。这导致速度提高了两个数量级,相对于TractCloud,甚至可以在没有GPU的临床工作站上进行推断。PETParc通过新型翻转不变嵌入或仅使用翻转作为数据扩充的一部分来解决流线方向缺失的问题。尽管加快了速度,但结果往往比以往的方法更好。论文接受后将公开代码和预训练模型。
论文及项目相关链接
PDF 8 pages, 2 pages references, 3 figures, 2 tables
Summary
全新大脑纤维追踪方法PETParc提出,采用并行高效追踪分割技术,基于transformer架构,随机分割全脑纤维图成子纤维图并行处理,相互作为全局背景,大幅提高处理速度,支持临床工作站无GPU推理。解决因流线方向缺失的问题,通过新型翻转不变嵌入或利用翻转进行数据增强实现。代码和预训练模型公开。
Key Takeaways
- PETParc是一种基于并行高效追踪分割技术的方法,旨在提高全脑纤维追踪图的分割效率。
- PETParc采用transformer架构,随机分割全脑纤维图为子纤维图,进行并行处理,相互作为全局背景。
- PETParc处理速度大幅提高,可达到TractCloud的两百倍速,且能在无GPU的临床工作站进行推理。
- PETParc解决了因流线方向缺失的问题,可以通过新型翻转不变嵌入或利用翻转进行数据增强实现。
- PETParc相对于之前的方法,结果往往更好。
- PETParc公开代码和预训练模型供用户使用和进一步研究。
点此查看论文截图

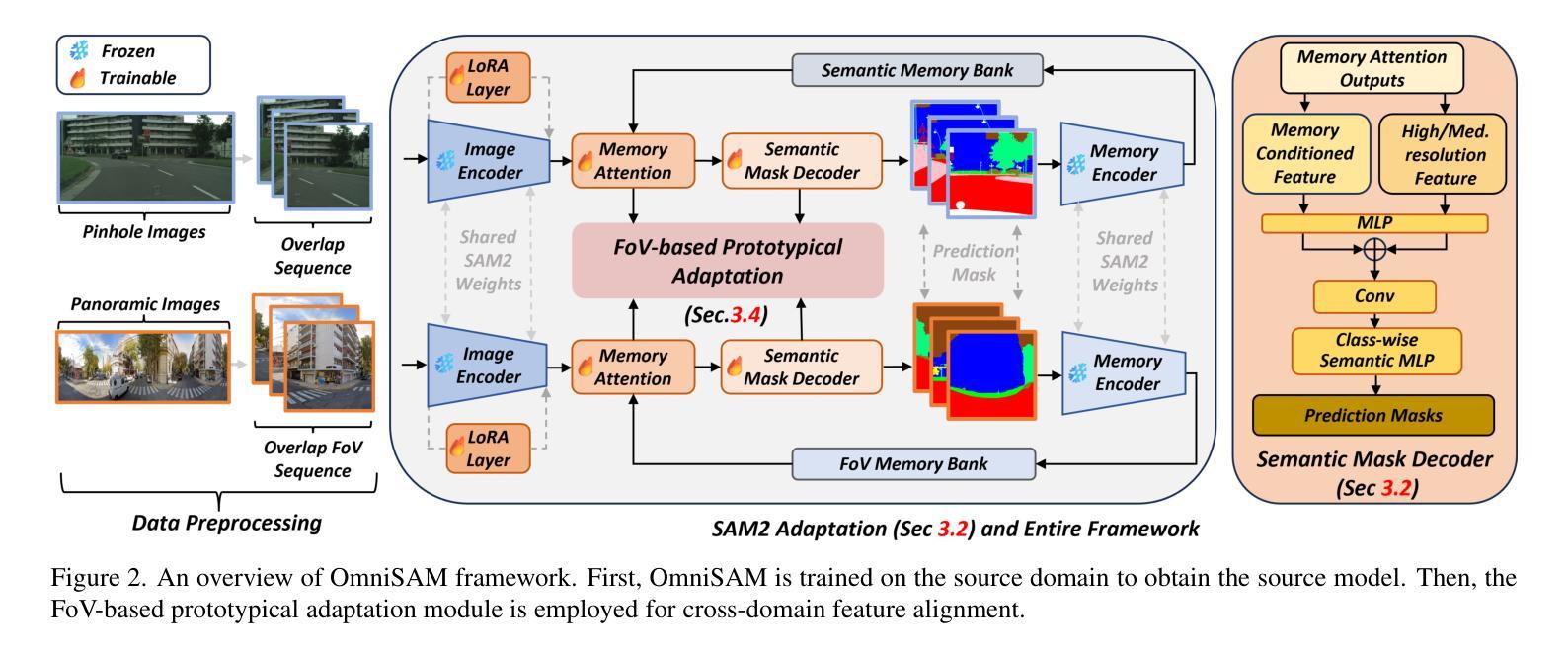
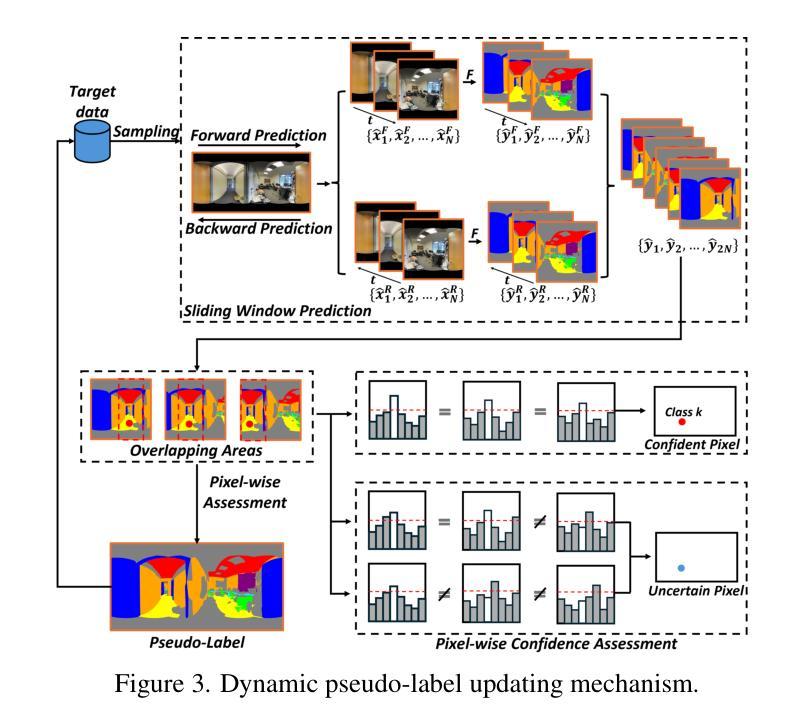
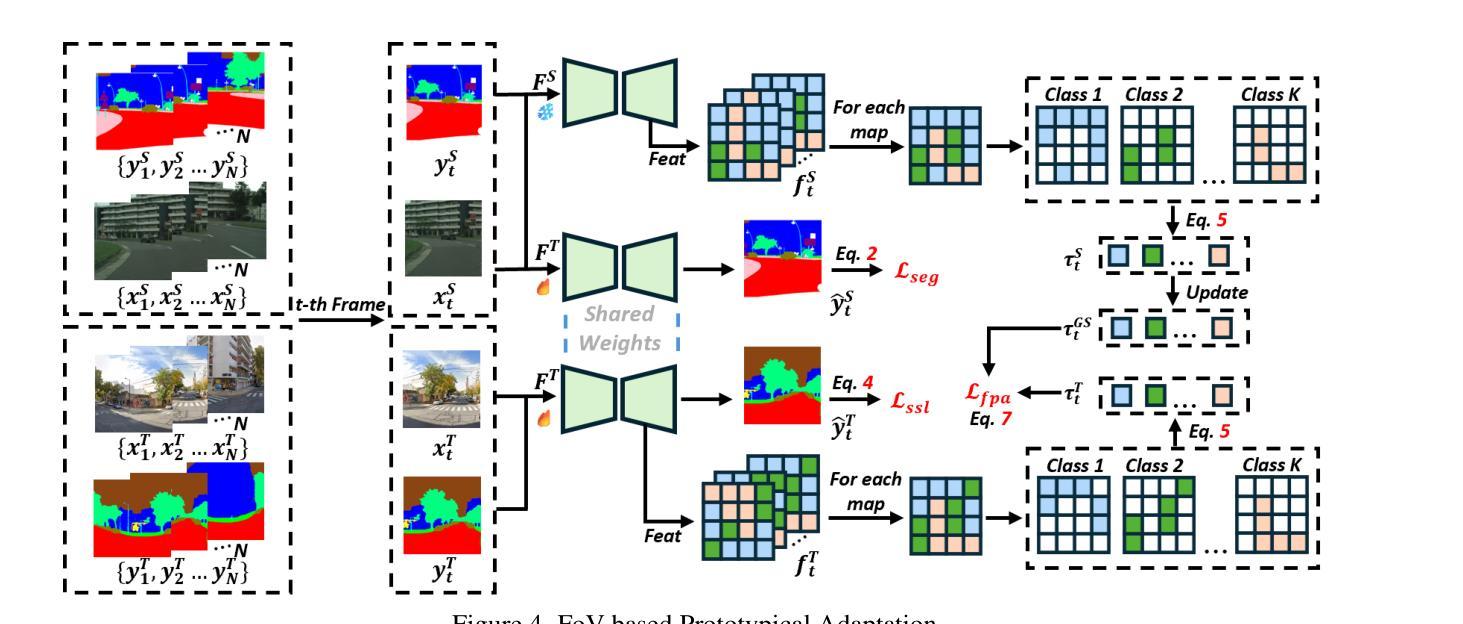
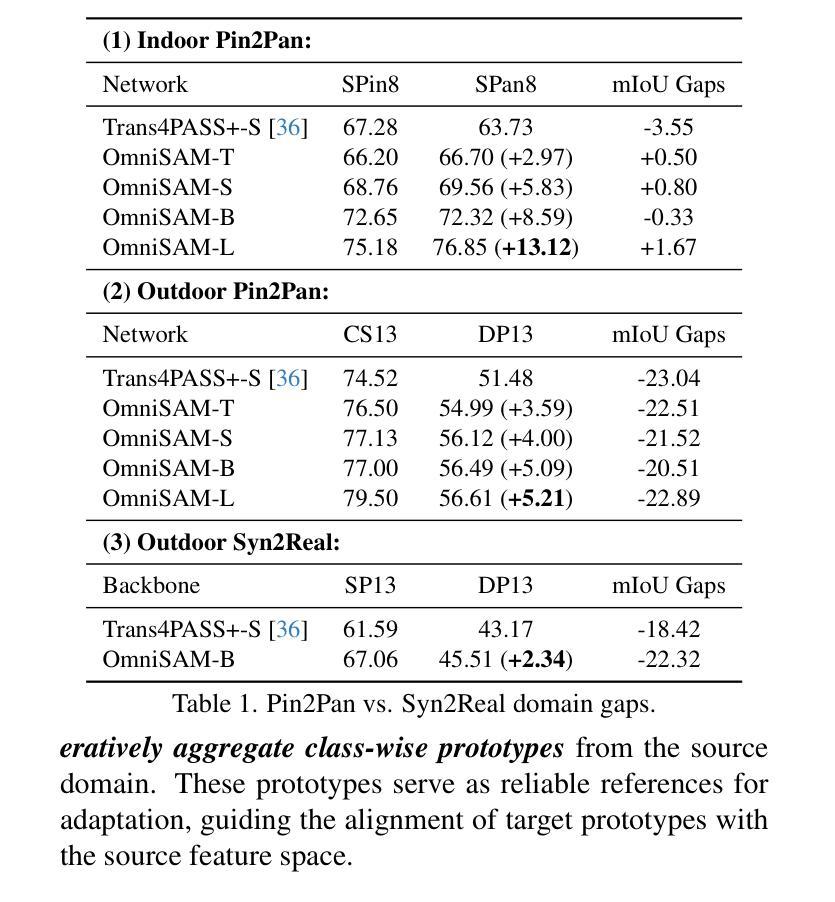
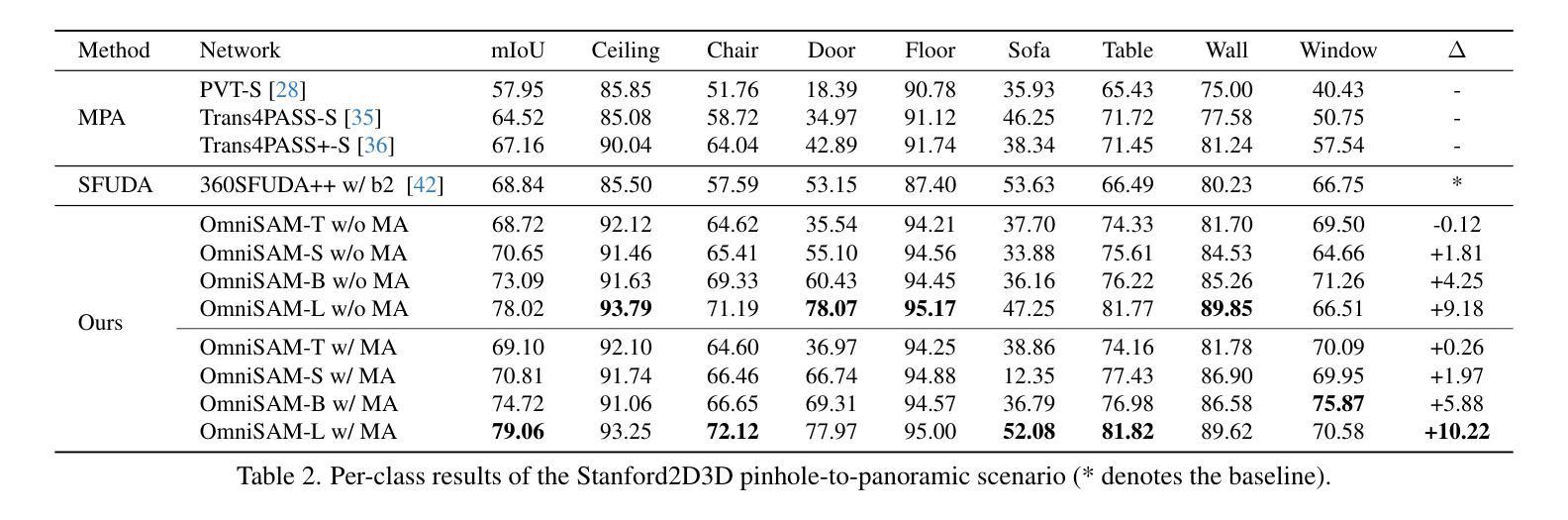
OmniSAM: Omnidirectional Segment Anything Model for UDA in Panoramic Semantic Segmentation
Authors:Ding Zhong, Xu Zheng, Chenfei Liao, Yuanhuiyi Lyu, Jialei Chen, Shengyang Wu, Linfeng Zhang, Xuming Hu
Segment Anything Model 2 (SAM2) has emerged as a strong base model in various pinhole imaging segmentation tasks. However, when applying it to $360^\circ$ domain, the significant field-of-view (FoV) gap between pinhole ($70^\circ \times 70^\circ$) and panoramic images ($180^\circ \times 360^\circ$) poses unique challenges. Two major concerns for this application includes 1) inevitable distortion and object deformation brought by the large FoV disparity between domains; 2) the lack of pixel-level semantic understanding that the original SAM2 cannot provide. To address these issues, we propose a novel OmniSAM framework, which makes the first attempt to apply SAM2 for panoramic semantic segmentation. Specifically, to bridge the first gap, OmniSAM first divides the panorama into sequences of patches. These patches are then treated as image sequences in similar manners as in video segmentation tasks. We then leverage the SAM2’s memory mechanism to extract cross-patch correspondences that embeds the cross-FoV dependencies, improving feature continuity and the prediction consistency along mask boundaries. For the second gap, OmniSAM fine-tunes the pretrained image encoder and reutilize the mask decoder for semantic prediction. An FoV-based prototypical adaptation module with dynamic pseudo label update mechanism is also introduced to facilitate the alignment of memory and backbone features, thereby improving model generalization ability across different sizes of source models. Extensive experimental results demonstrate that OmniSAM outperforms the state-of-the-art methods by large margins, e.g., 79.06% (+10.22%) on SPin8-to-SPan8, 62.46% (+6.58%) on CS13-to-DP13.
Segment Anything Model 2(SAM2)在各种针孔成像分割任务中已成为强大的基础模型。然而,将其应用于$360^\circ$域时,针孔($70^\circ \times 70^\circ$)和全景图像($180^\circ \times 360^\circ$)之间的视场(FoV)差距巨大,带来了独特的挑战。对此应用的主要担忧包括:1)由域之间的大视场差异带来的不可避免的形状扭曲和物体变形;2)原始SAM2无法提供像素级的语义理解。为了解决这些问题,我们提出了全新的OmniSAM框架,它首次尝试将SAM2应用于全景语义分割。具体来说,为了弥补第一个差距,OmniSAM首先把全景分成一系列的补丁。这些补丁然后以类似于视频分割任务的方式作为图像序列进行处理。然后,我们利用SAM2的记忆机制来提取跨补丁对应关系,这些对应关系嵌入了跨视场的依赖关系,提高了特征连续性和掩膜边界的预测一致性。对于第二个差距,OmniSAM对预训练图像编码器进行微调,并重新使用掩膜解码器进行语义预测。还引入了一个基于视场的原型自适应模块,带有动态伪标签更新机制,以促进记忆和主干特征的对齐,从而提高模型在不同大小源模型上的泛化能力。大量的实验结果证明,OmniSAM大大超越了最先进的方法,例如在SPin8-to-SPan8上达到79.06%(+10.22%),在CS13-to-DP13上达到62.46%(+6.58%)。
论文及项目相关链接
Summary
全景图像语义分割中,SAM2模型存在视野(FoV)差距和像素级语义理解不足的问题。为此,提出OmniSAM框架,通过分割全景图像为序列补丁,利用SAM2的记忆机制提取跨补丁对应关系,改善特征连续性和预测一致性。同时,微调预训练图像编码器并重新利用掩膜解码器进行语义预测,引入基于视野的原型适应模块和动态伪标签更新机制,提高模型在不同尺寸源模型上的泛化能力。实验结果证明OmniSAM大幅优于现有方法。
Key Takeaways
- SAM2模型在全景成像语义分割中面临视野差距和像素级语义理解不足的挑战。
- OmniSAM框架提出将全景图像分割成序列补丁,以缩小视野差距。
- OmniSAM利用SAM2的记忆机制提取跨补丁对应关系,改善特征连续性和预测一致性。
- OmniSAM通过微调预训练图像编码器和重新利用掩膜解码器来应对像素级语义理解不足的问题。
- OmniSAM引入基于视野的原型适应模块,提高模型在不同尺寸源模型上的泛化能力。
- 动态伪标签更新机制被用于促进记忆和主干特征的对齐。
- 实验结果显示OmniSAM大幅优于现有方法,如在SPin8-to-SPan8上达到79.06%(+10.22%),在CS13-to-DP13上达到62.46%(+6.58%)。
点此查看论文截图

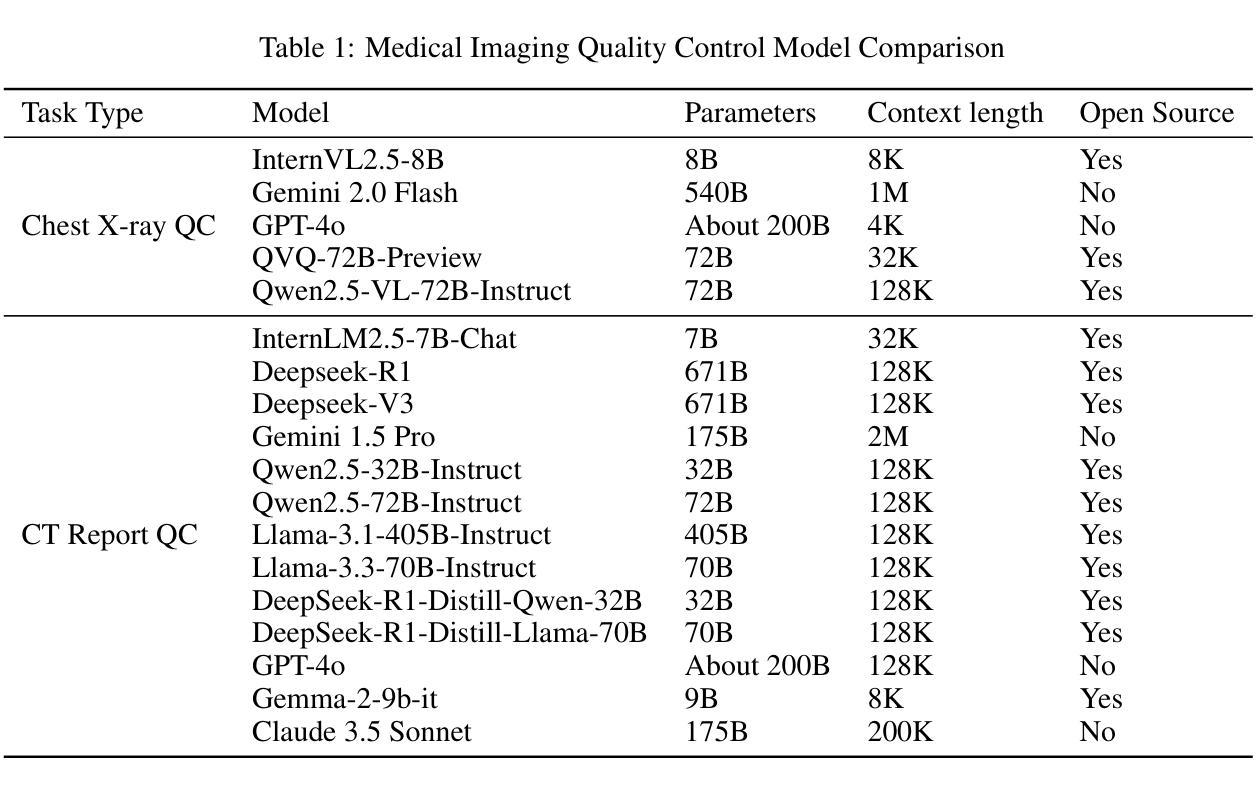
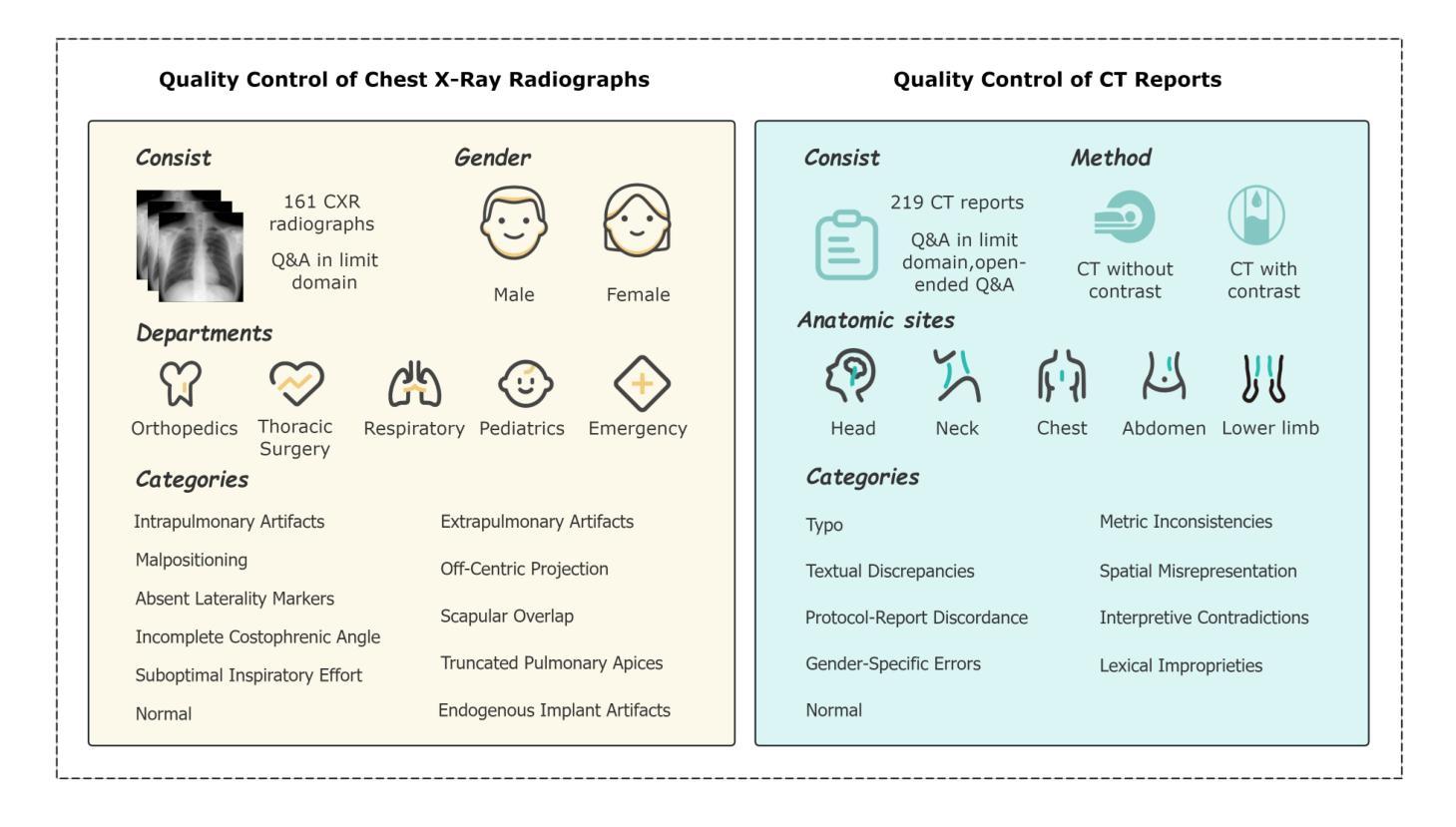
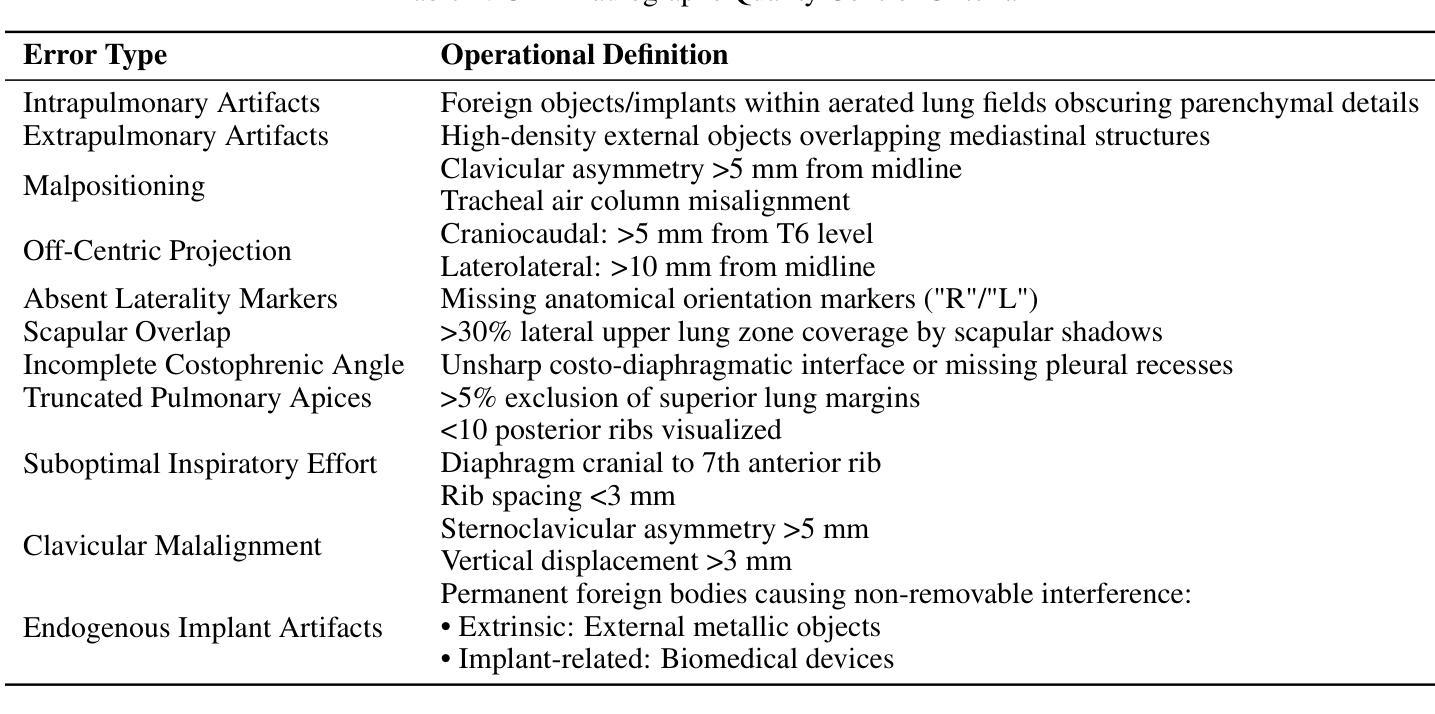
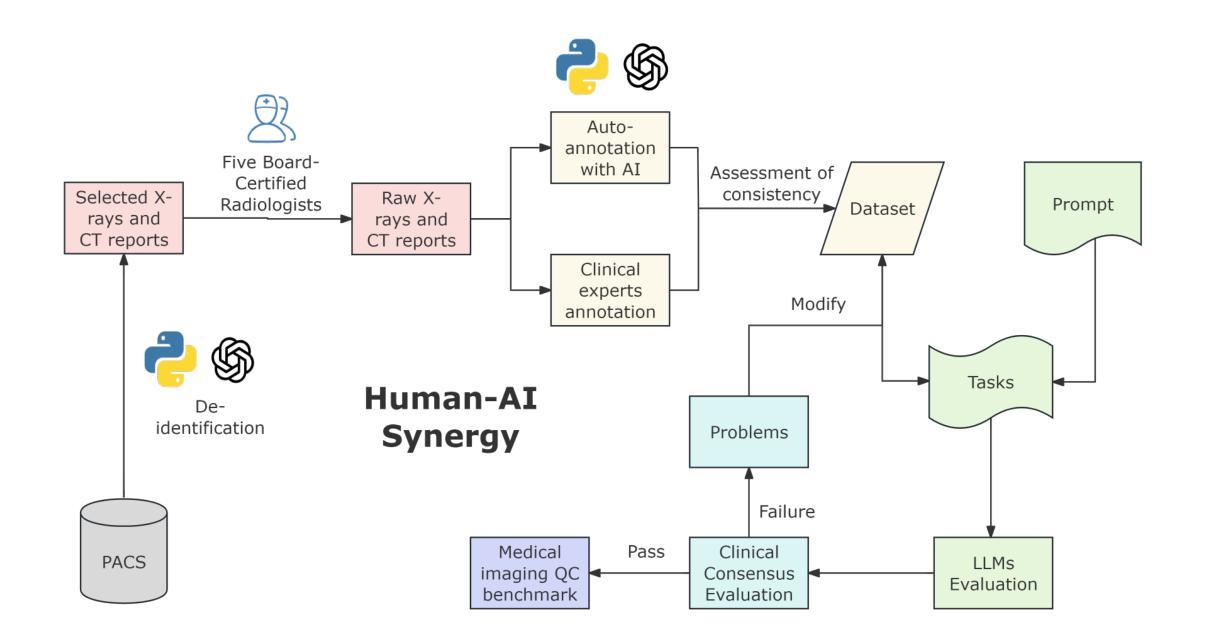
Multimodal Human-AI Synergy for Medical Imaging Quality Control: A Hybrid Intelligence Framework with Adaptive Dataset Curation and Closed-Loop Evaluation
Authors:Zhi Qin, Qianhui Gui, Mouxiao Bian, Rui Wang, Hong Ge, Dandan Yao, Ziying Sun, Yuan Zhao, Yu Zhang, Hui Shi, Dongdong Wang, Chenxin Song, Shenghong Ju, Lihao Liu, Junjun He, Jie Xu, Yuan-Cheng Wang
Medical imaging quality control (QC) is essential for accurate diagnosis, yet traditional QC methods remain labor-intensive and subjective. To address this challenge, in this study, we establish a standardized dataset and evaluation framework for medical imaging QC, systematically assessing large language models (LLMs) in image quality assessment and report standardization. Specifically, we first constructed and anonymized a dataset of 161 chest X-ray (CXR) radiographs and 219 CT reports for evaluation. Then, multiple LLMs, including Gemini 2.0-Flash, GPT-4o, and DeepSeek-R1, were evaluated based on recall, precision, and F1 score to detect technical errors and inconsistencies. Experimental results show that Gemini 2.0-Flash achieved a Macro F1 score of 90 in CXR tasks, demonstrating strong generalization but limited fine-grained performance. DeepSeek-R1 excelled in CT report auditing with a 62.23% recall rate, outperforming other models. However, its distilled variants performed poorly, while InternLM2.5-7B-chat exhibited the highest additional discovery rate, indicating broader but less precise error detection. These findings highlight the potential of LLMs in medical imaging QC, with DeepSeek-R1 and Gemini 2.0-Flash demonstrating superior performance.
医学影像质量控制(QC)对于准确诊断至关重要,然而传统的QC方法仍然劳动强度高且主观。为了应对这一挑战,本研究建立了医学影像QC的标准数据集和评估框架,对图像质量评估和报告标准化的大型语言模型(LLMs)进行了系统评估。具体来说,我们首先构建并匿名化了一个包含161张胸部X射线(CXR)放射照片和219份CT报告的数据集用于评估。然后,基于召回率、精确度和F1分数,对包括Gemini 2.0-Flash、GPT-4o和DeepSeek-R1在内的多个LLMs进行了技术错误和不一致性的检测评估。实验结果表明,Gemini 2.0-Flash在CXR任务中获得了90的宏观F1分数,表现出较强的泛化能力但精细粒度性能有限。DeepSeek-R1在CT报告审核方面表现出色,召回率达到62.23%,优于其他模型。然而,其蒸馏变体表现不佳,而InternLM2.5-7B-chat表现出最高的额外发现率,表明其错误检测范围更广但精确度较低。这些发现突显了LLMs在医学影像QC中的潜力,其中DeepSeek-R1和Gemini 2.0-Flash表现出卓越的性能。
论文及项目相关链接
Summary
本文研究了医学成像质量控制(QC)的重要性及其面临的挑战。为解决这个问题,研究团队建立了一套标准化的数据集和评估框架,用于评估大型语言模型在医学图像质量评估和报告标准化方面的性能。通过构建匿名数据集和测试多种大型语言模型,发现某些模型在特定任务上表现出色,为医学成像QC提供了新的可能性。
Key Takeaways
- 医学成像质量控制对准确诊断至关重要,但传统方法劳动强度大、主观性强。
- 研究建立了一套标准化的数据集和评估框架,用于医学成像QC。
- 大型语言模型在图像质量评估和报告标准化方面进行了系统评估。
- Gemini 2.0-Flash在CXR任务中表现出强泛化能力,但精细粒度性能有限。
- DeepSeek-R1在CT报告审核方面表现出色,具有较高的召回率。
- InternLM2.5-7B-chat具有更广泛的错误检测能力,但精确性较低。
点此查看论文截图

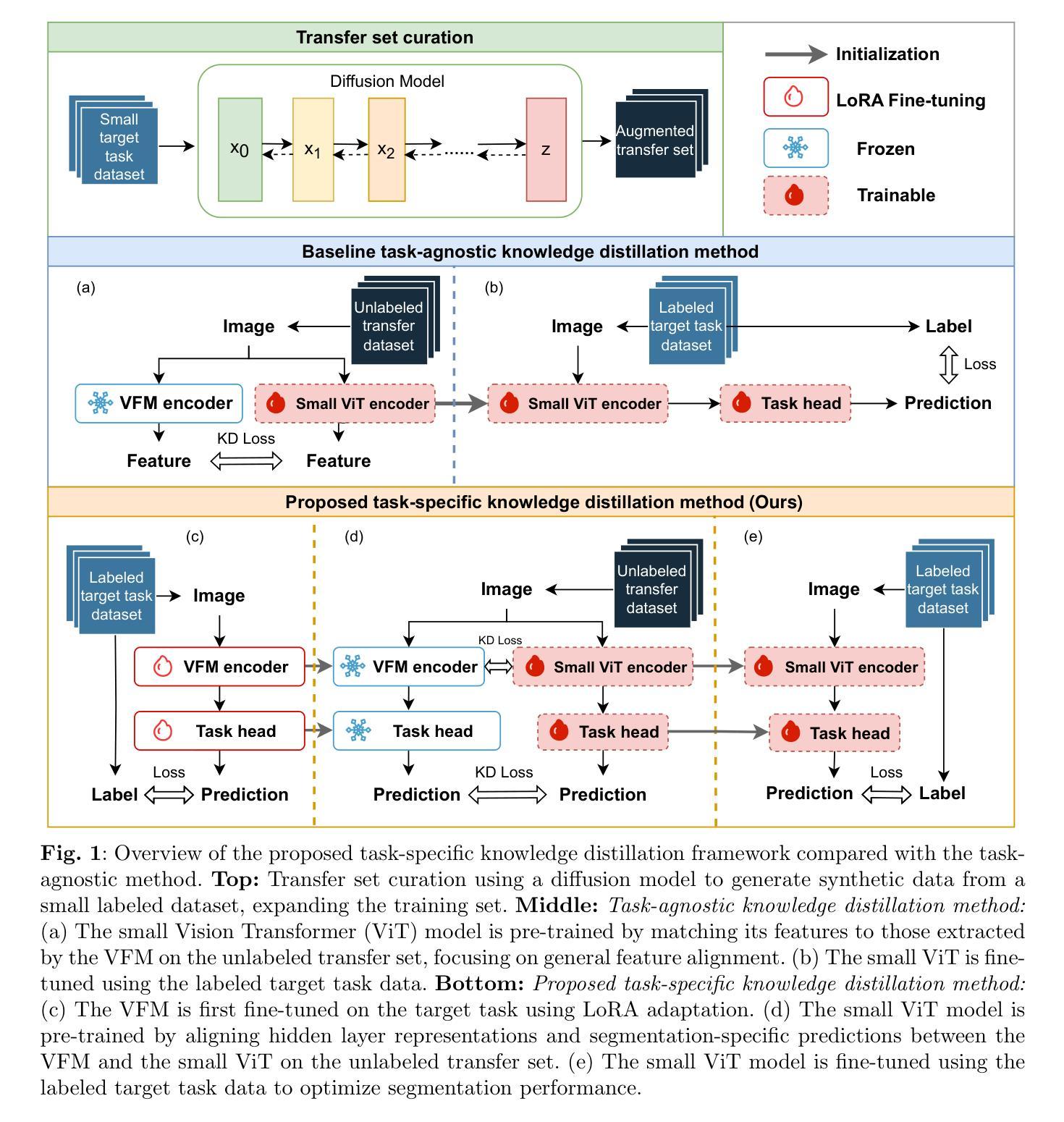
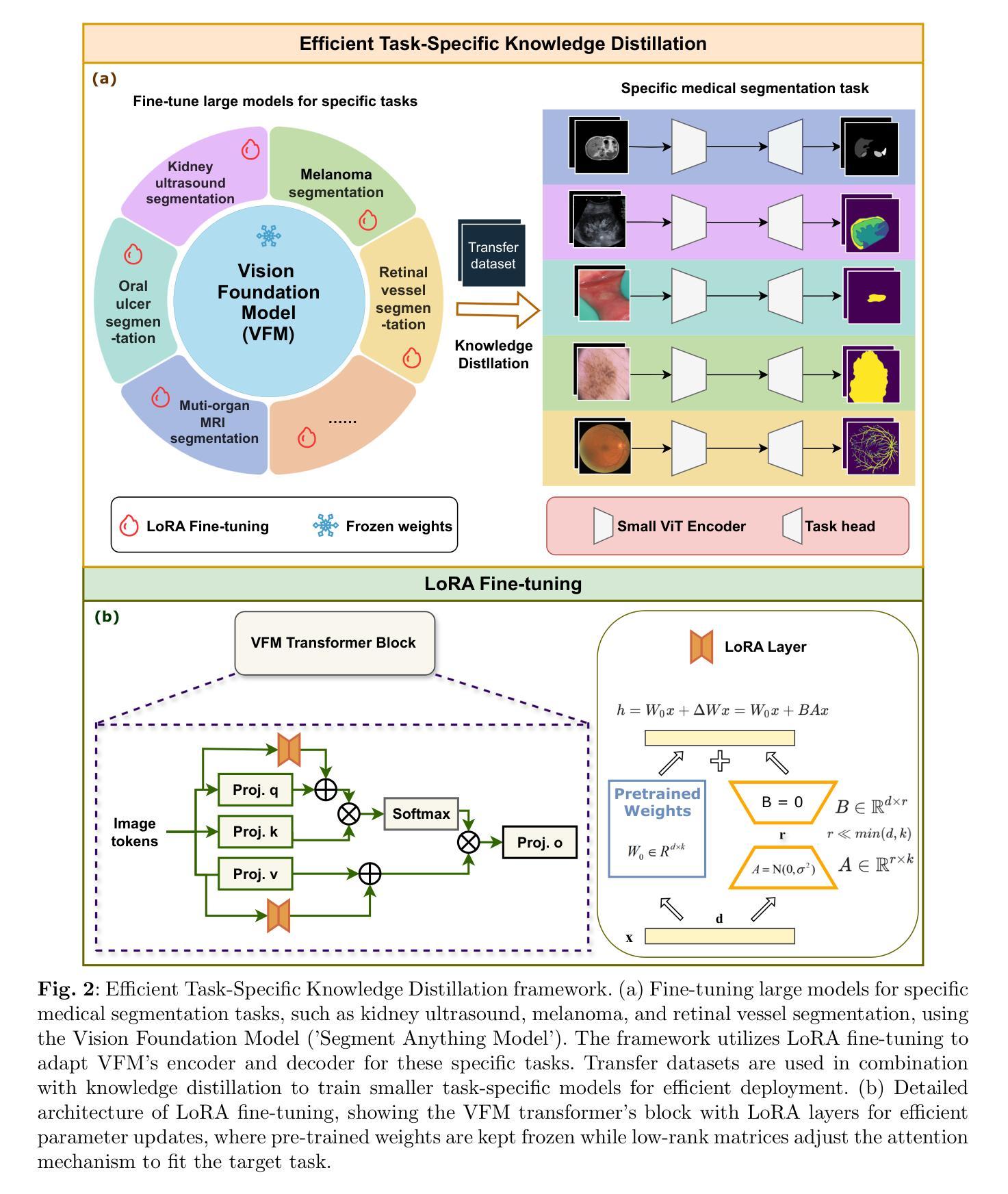
Task-Specific Knowledge Distillation from the Vision Foundation Model for Enhanced Medical Image Segmentation
Authors:Pengchen Liang, Haishan Huang, Bin Pu, Jianguo Chen, Xiang Hua, Jing Zhang, Weibo Ma, Zhuangzhuang Chen, Yiwei Li, Qing Chang
Large-scale pre-trained models, such as Vision Foundation Models (VFMs), have demonstrated impressive performance across various downstream tasks by transferring generalized knowledge, especially when target data is limited. However, their high computational cost and the domain gap between natural and medical images limit their practical application in medical segmentation tasks. Motivated by this, we pose the following important question: “How can we effectively utilize the knowledge of large pre-trained VFMs to train a small, task-specific model for medical image segmentation when training data is limited?” To address this problem, we propose a novel and generalizable task-specific knowledge distillation framework. Our method fine-tunes the VFM on the target segmentation task to capture task-specific features before distilling the knowledge to smaller models, leveraging Low-Rank Adaptation (LoRA) to reduce the computational cost of fine-tuning. Additionally, we incorporate synthetic data generated by diffusion models to augment the transfer set, enhancing model performance in data-limited scenarios. Experimental results across five medical image datasets demonstrate that our method consistently outperforms task-agnostic knowledge distillation and self-supervised pretraining approaches like MoCo v3 and Masked Autoencoders (MAE). For example, on the KidneyUS dataset, our method achieved a 28% higher Dice score than task-agnostic KD using 80 labeled samples for fine-tuning. On the CHAOS dataset, it achieved an 11% improvement over MAE with 100 labeled samples. These results underscore the potential of task-specific knowledge distillation to train accurate, efficient models for medical image segmentation in data-constrained settings.
大规模预训练模型,如视觉基础模型(VFMs),通过迁移通用知识,在各种下游任务中展现了令人印象深刻的性能,尤其是在目标数据有限的情况下。然而,它们的高计算成本和自然图像与医学图像之间的领域差距限制了它们在医学分割任务中的实际应用。因此,我们提出了以下重要问题:“当训练数据有限时,如何有效利用大规模预训练VFMs的知识来训练一个小而特定的医学图像分割模型?”为了解决这一问题,我们提出了一种新型且通用的任务特定知识蒸馏框架。我们的方法首先在目标分割任务上微调VFM,以捕获任务特定特征,然后将知识蒸馏到较小的模型,并利用低阶适应(LoRA)来降低微调的计算成本。此外,我们结合扩散模型生成的合成数据来增强转移集,提高模型在数据有限场景中的性能。在五个医学图像数据集上的实验结果表明,我们的方法一直优于任务无关的知识蒸馏和自监督预训练方法,如MoCo v3和掩码自动编码器(MAE)。例如,在KidneyUS数据集上,使用80个标记样本进行微调时,我们的方法得到的Dice得分比任务无关KD高出28%。在CHAOS数据集上,使用100个标记样本时,它比MAE提高了11%。这些结果突出了任务特定知识蒸馏在数据受限环境中训练医学图像分割的准确、高效模型的潜力。
论文及项目相关链接
PDF 29 pages, 10 figures, 16 tables
摘要
利用大规模预训练模型(如视觉基础模型)的通用知识,提出一种针对医疗图像分割任务的有效知识蒸馏框架。该框架在目标分割任务上微调预训练模型,捕获任务特定特征,并将知识蒸馏到小型模型中。通过低秩适应(LoRA)降低微调的计算成本,同时使用扩散模型生成的合成数据增强转移集。实验结果表明,该方法在五个医疗图像数据集上的性能均优于任务无关的知识蒸馏和自我监督预训练方法,如MoCo v3和掩码自动编码器(MAE)。
关键见解
- 大规模预训练模型,如视觉基础模型(VFMs),在迁移学习时表现出优异的性能,尤其在目标数据有限的情况下。
- 针对医疗图像分割任务,利用预训练模型的知识是关键。
- 提出一种新型的任务特定知识蒸馏框架,通过微调预训练模型来捕获任务特定特征,并将知识传递给小型模型。
- 使用低秩适应(LoRA)降低微调的计算成本,提高模型的实用性。
- 结合扩散模型生成的合成数据增强转移集,提高模型在数据有限场景中的性能。
- 在多个医疗图像数据集上的实验结果表明,该方法优于任务无关的知识蒸馏和自我监督预训练方法。
- 在肾脏US和CHAOS数据集上的实验结果显示,该方法在有限的标注数据下实现了显著的性能提升。
点此查看论文截图

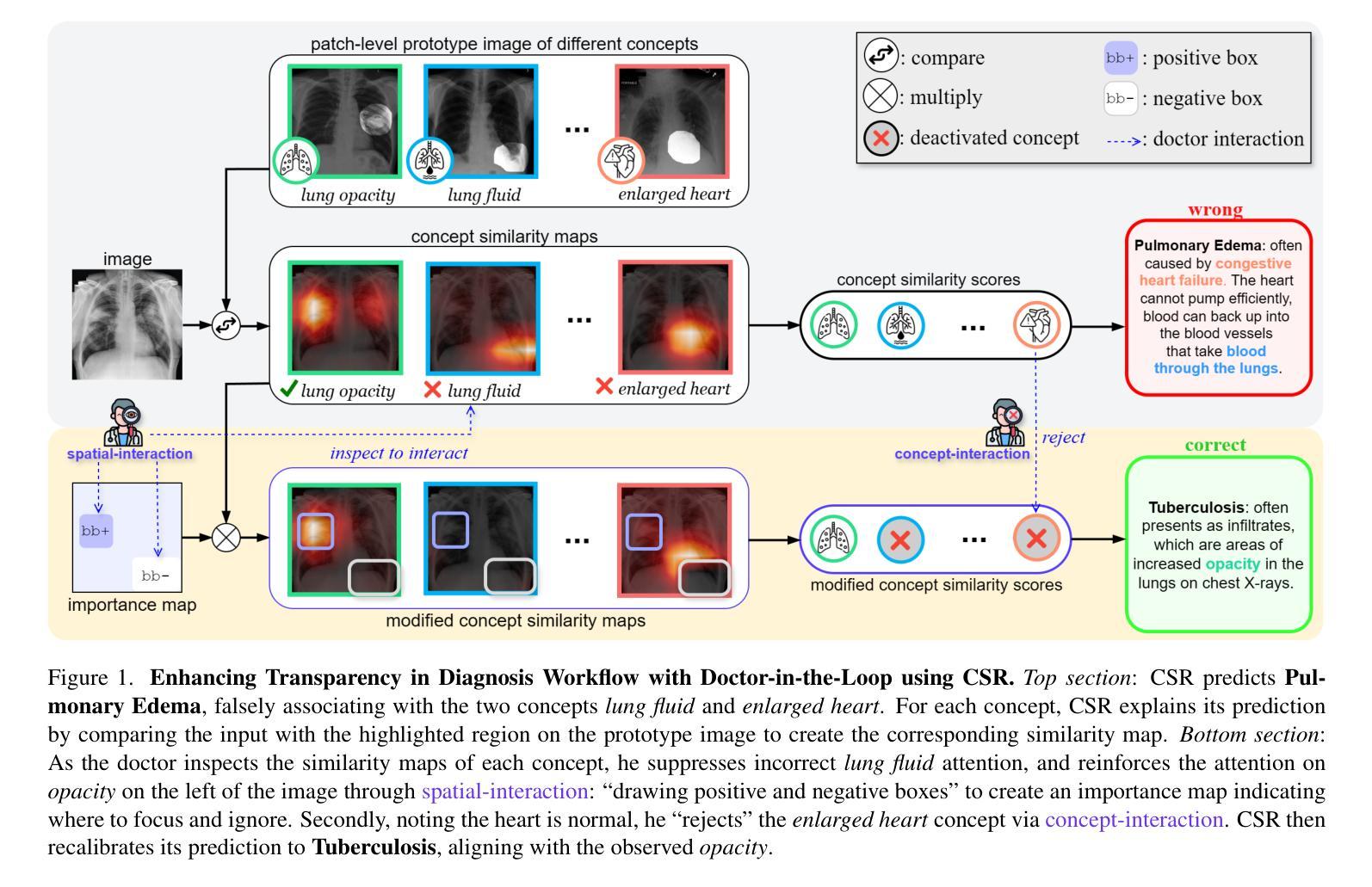
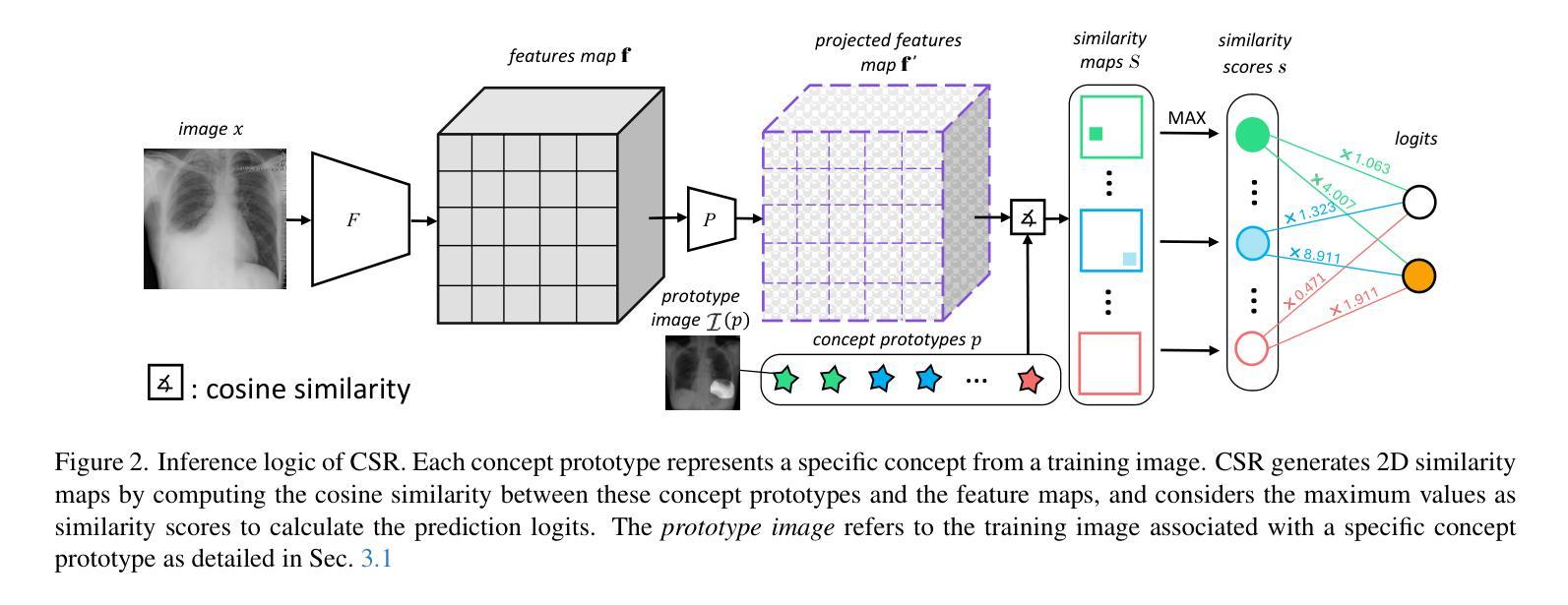
Interactive Medical Image Analysis with Concept-based Similarity Reasoning
Authors:Ta Duc Huy, Sen Kim Tran, Phan Nguyen, Nguyen Hoang Tran, Tran Bao Sam, Anton van den Hengel, Zhibin Liao, Johan W. Verjans, Minh-Son To, Vu Minh Hieu Phan
The ability to interpret and intervene model decisions is important for the adoption of computer-aided diagnosis methods in clinical workflows. Recent concept-based methods link the model predictions with interpretable concepts and modify their activation scores to interact with the model. However, these concepts are at the image level, which hinders the model from pinpointing the exact patches the concepts are activated. Alternatively, prototype-based methods learn representations from training image patches and compare these with test image patches, using the similarity scores for final class prediction. However, interpreting the underlying concepts of these patches can be challenging and often necessitates post-hoc guesswork. To address this issue, this paper introduces the novel Concept-based Similarity Reasoning network (CSR), which offers (i) patch-level prototype with intrinsic concept interpretation, and (ii) spatial interactivity. First, the proposed CSR provides localized explanation by grounding prototypes of each concept on image regions. Second, our model introduces novel spatial-level interaction, allowing doctors to engage directly with specific image areas, making it an intuitive and transparent tool for medical imaging. CSR improves upon prior state-of-the-art interpretable methods by up to 4.5% across three biomedical datasets. Our code is released at https://github.com/tadeephuy/InteractCSR.
解释和干预模型决策的能力对于在临床工作流程中采用计算机辅助诊断方法至关重要。最近的概念级方法将模型预测与可解释的概念联系起来,并修改其激活分数以与模型进行交互。然而,这些概念仅限于图像层面,阻碍了模型精确识别概念被激活的确切区域。另一种基于原型的方法从训练图像块中学习表示,并将其与测试图像块进行比较,使用相似度分数进行最终的类别预测。然而,解释这些图像块的潜在概念可能具有挑战性,通常需要事后猜测。为了解决这个问题,本文引入了新型的概念相似性推理网络(CSR),它提供了(i)图像块级的原型与内在概念解释,(ii)空间交互性。首先,所提出的CSR通过在图像区域上为每个概念提供原型来提供局部解释。其次,我们的模型引入了新型的空间级别交互,使医生能够直接与特定的图像区域进行交互,使其成为医学影像的直观和透明工具。CSR在三个生物医学数据集上较之前的最先进的可解释方法提高了高达4.5%。我们的代码已发布在https://github.com/tadeephuy/InteractCSR。
论文及项目相关链接
PDF Accepted CVPR2025
Summary
医学图像领域中的新概念——基于相似性的概念推理网络(CSR)。CSR可实现:(一)区域化的概念解释,即定位到图像区域的概念原型;(二)空间互动,使得医生可直观地直接与特定图像区域互动。该方法提高了医学图像诊断的准确性和透明度。
Key Takeaways
- 计算机辅助诊断方法的采用需要解释和干预模型决策的能力。
- 当前概念级方法通过连接模型预测与可解释概念来优化激活得分,从而与模型进行交互,但这种交互方法不能精确地确定概念激活的具体区域。
- 基于原型的方法通过学习从训练图像补丁表示并与测试图像补丁进行比较来预测最终类别,但解释这些补丁的潜在概念可能具有挑战性,并需要事后猜测。
- 新提出的CSR网络解决了上述问题,通过提供区域化的概念解释和空间互动功能,实现了更精确的模型预测和医生互动。
- CSR网络在三个生物医学数据集上较之前的最先进的可解释方法提高了高达4.5%。
- CSR网络通过定位概念原型在图像区域中的位置提供本地化解释。
点此查看论文截图

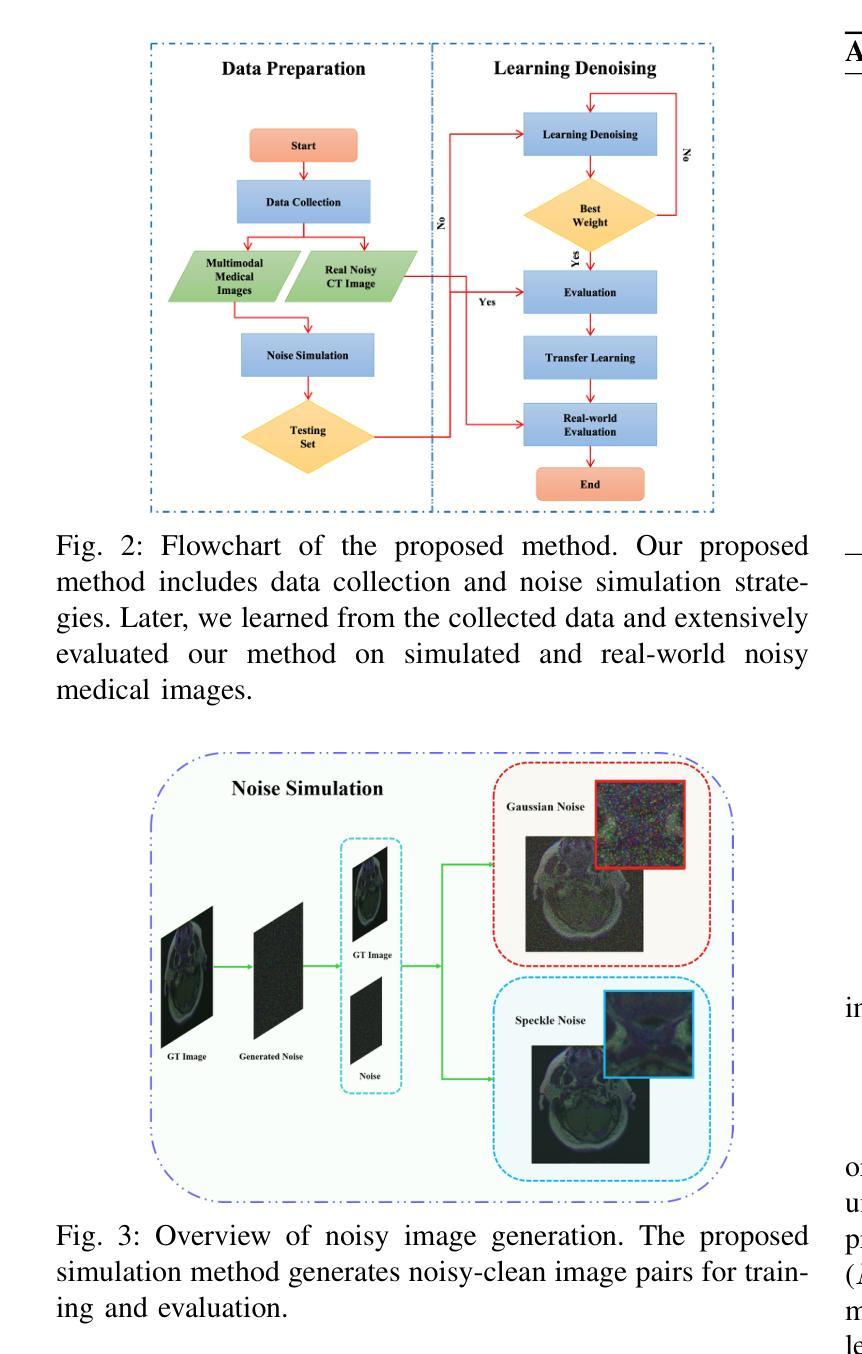
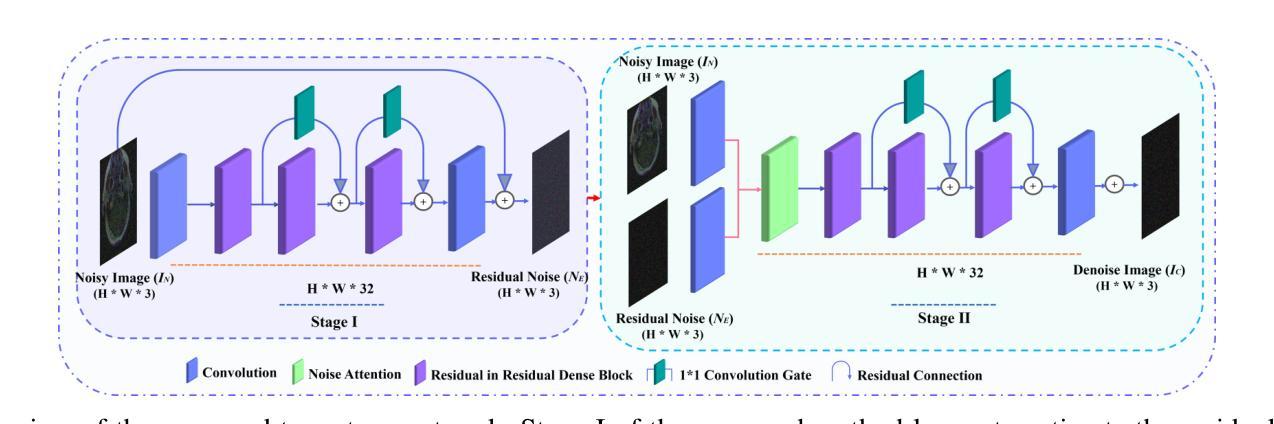
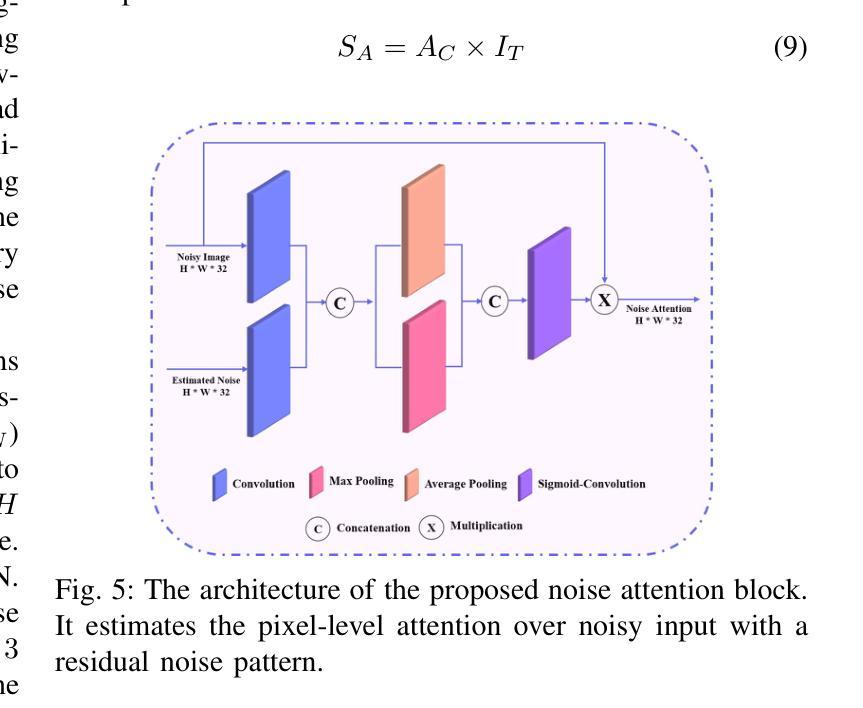
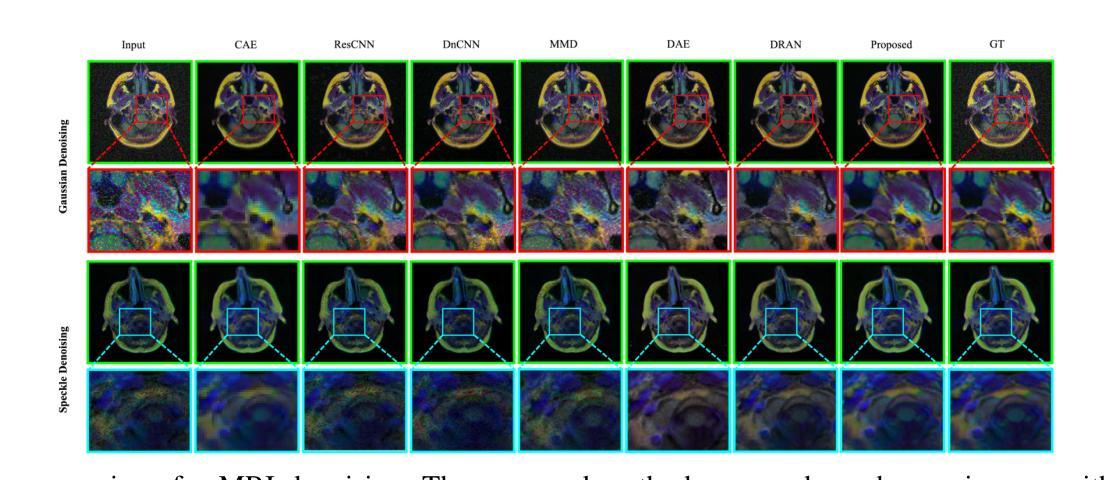
Two-stage Deep Denoising with Self-guided Noise Attention for Multimodal Medical Images
Authors:S M A Sharif, Rizwan Ali Naqvi, Woong-Kee Loh
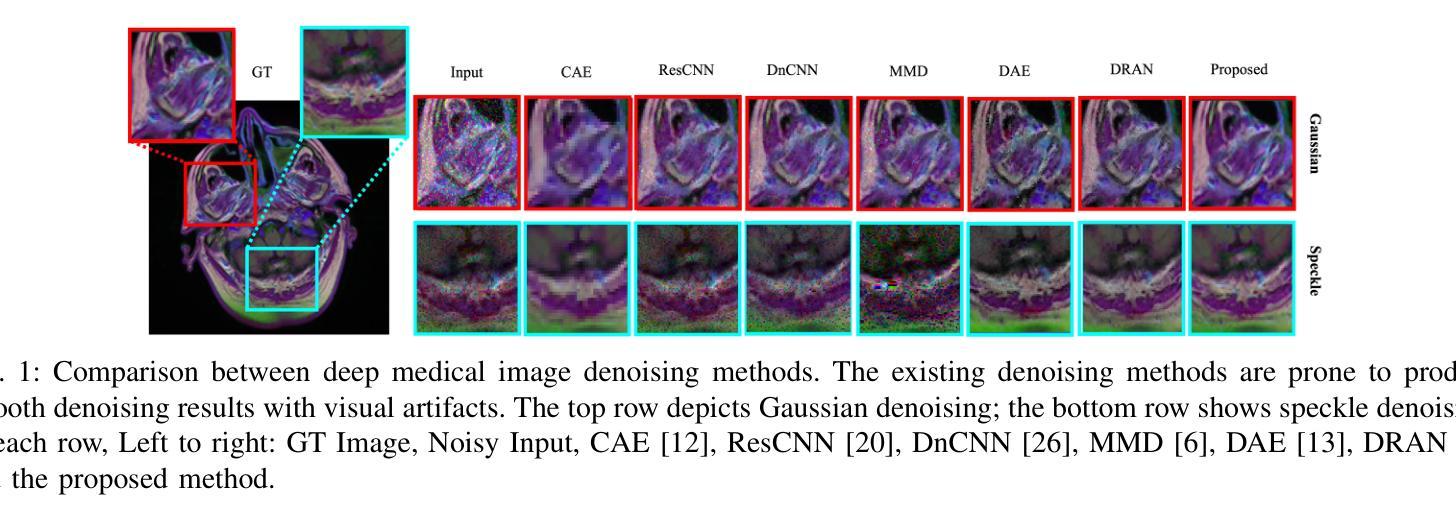
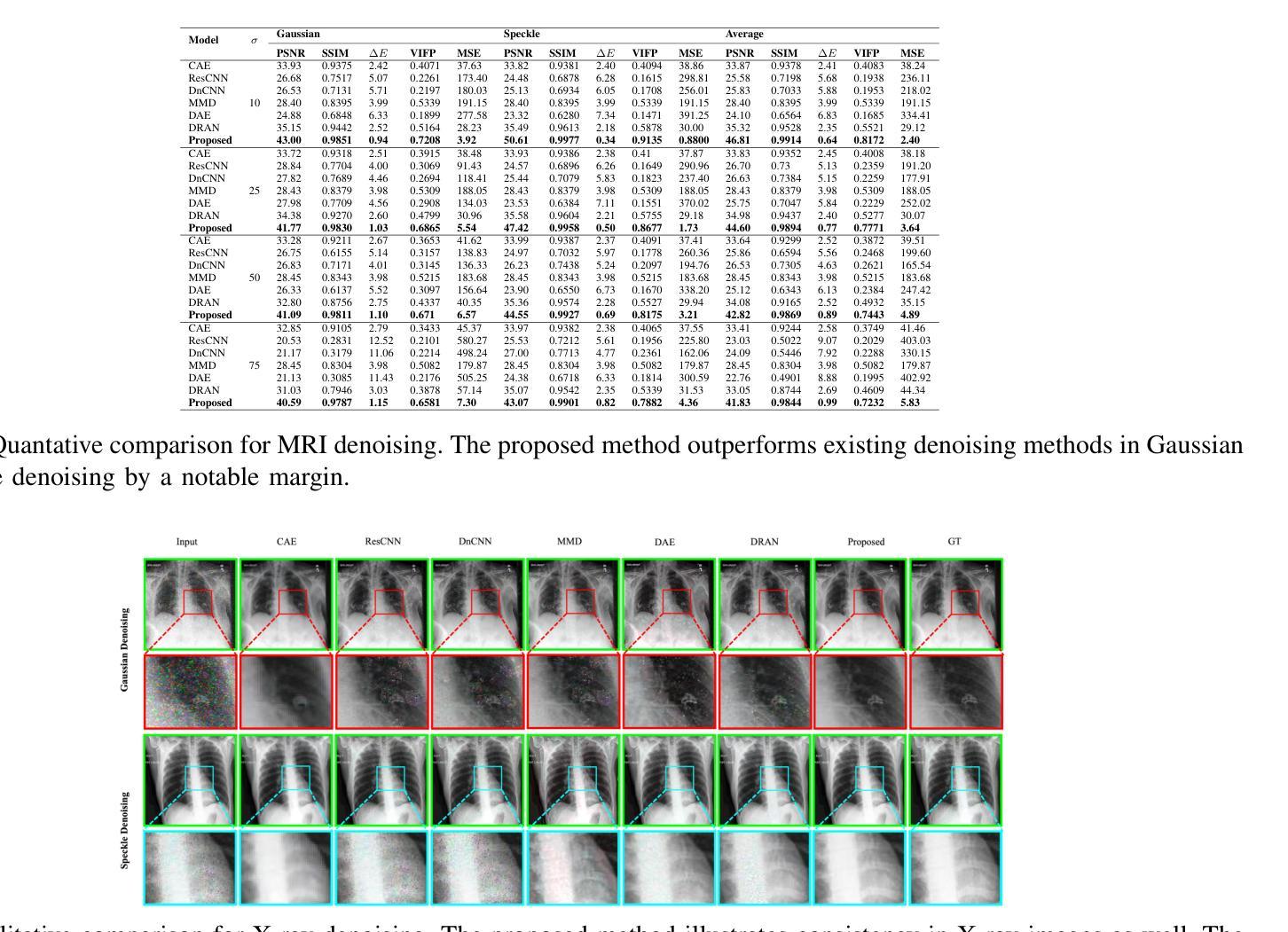
Medical image denoising is considered among the most challenging vision tasks. Despite the real-world implications, existing denoising methods have notable drawbacks as they often generate visual artifacts when applied to heterogeneous medical images. This study addresses the limitation of the contemporary denoising methods with an artificial intelligence (AI)-driven two-stage learning strategy. The proposed method learns to estimate the residual noise from the noisy images. Later, it incorporates a novel noise attention mechanism to correlate estimated residual noise with noisy inputs to perform denoising in a course-to-refine manner. This study also proposes to leverage a multi-modal learning strategy to generalize the denoising among medical image modalities and multiple noise patterns for widespread applications. The practicability of the proposed method has been evaluated with dense experiments. The experimental results demonstrated that the proposed method achieved state-of-the-art performance by significantly outperforming the existing medical image denoising methods in quantitative and qualitative comparisons. Overall, it illustrates a performance gain of 7.64 in Peak Signal-to-Noise Ratio (PSNR), 0.1021 in Structural Similarity Index (SSIM), 0.80 in DeltaE ($\Delta E$), 0.1855 in Visual Information Fidelity Pixel-wise (VIFP), and 18.54 in Mean Squared Error (MSE) metrics.
医学图像去噪被认为是视觉任务中最具挑战性的之一。尽管在实际应用中具有重要意义,现有的去噪方法在应用到异质医学图像时,往往会产生视觉伪影,存在明显的缺点。本研究通过人工智能(AI)驱动的两阶段学习策略来解决当前去噪方法的局限。所提出的方法学习估计噪声图像中的残余噪声。然后,它引入了一种新型的噪声注意力机制,将估计的残余噪声与带噪声的输入相关联,以从粗到细的方式进行去噪。本研究还提出利用多模式学习策略,以推广不同医学图像模式和多噪声模式的去噪应用。所提出方法的实用性已经通过大量实验进行了评估。实验结果表明,所提出的方法在定量和定性比较中显著优于现有医学图像去噪方法,达到了最先进的性能。总体而言,它在峰值信噪比(PSNR)上获得了7.64的性能提升,结构相似性指数(SSIM)提高了0.1021,DeltaE(ΔE)降低了0.80,视觉信息保真度像素级(VIFP)提高了0.1855,均方误差(MSE)指标减少了18.54。
论文及项目相关链接
PDF IEEE Transactions on Radiation and Plasma Medical Sciences (2024)
Summary
本文提出一种基于人工智能的两阶段学习策略,用于解决当前医学图像去噪方法的局限性。该方法能够估计噪声图像中的残余噪声,并结合噪声关注机制,以粗略到精细的方式进行去噪。同时,提出多模态学习策略,使去噪方法能够在不同医学图像模式和多噪声模式中得到广泛应用。实验证明,该方法在定量和定性比较中均优于现有医学图像去噪方法,实现了卓越的性能。
Key Takeaways
- 医学图像去噪是极具挑战性的视觉任务。
- 现有去噪方法在应用于异质医学图像时会产生视觉伪影。
- 本研究采用人工智能两阶段学习策略来解决现有去噪方法的局限性。
- 所提出的方法能够估计噪声图像中的残余噪声。
- 结合噪声关注机制,以粗略到精细的方式进行去噪。
- 提出多模态学习策略,提高去噪方法的泛化能力,适用于多种医学图像模式和多噪声模式。
点此查看论文截图

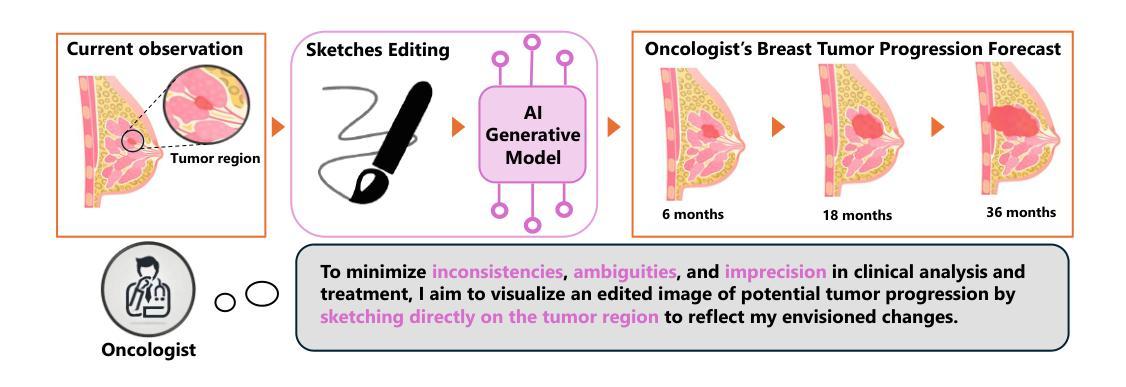
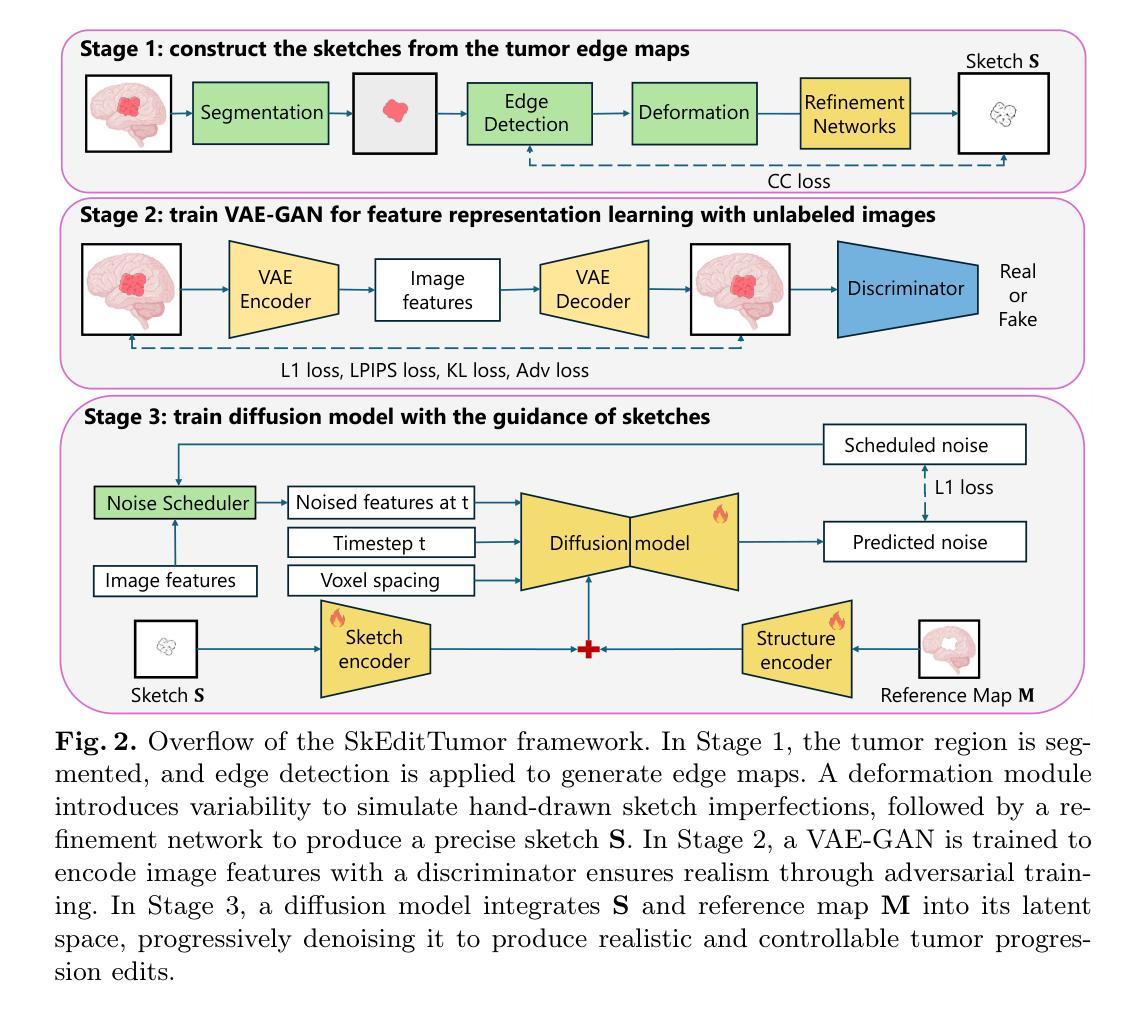
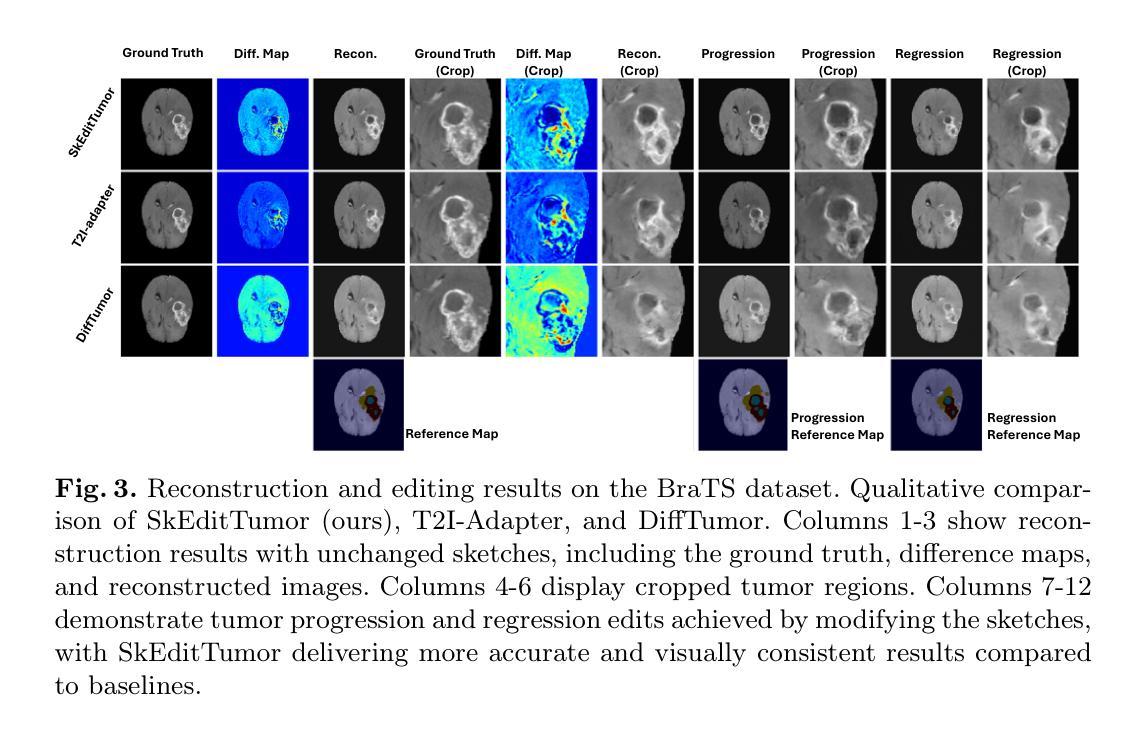
Interactive Tumor Progression Modeling via Sketch-Based Image Editing
Authors:Gexin Huang, Ruinan Jin, Yucheng Tang, Can Zhao, Tatsuya Harada, Xiaoxiao Li, Gu Lin
Accurately visualizing and editing tumor progression in medical imaging is crucial for diagnosis, treatment planning, and clinical communication. To address the challenges of subjectivity and limited precision in existing methods, we propose SkEditTumor, a sketch-based diffusion model for controllable tumor progression editing. By leveraging sketches as structural priors, our method enables precise modifications of tumor regions while maintaining structural integrity and visual realism. We evaluate SkEditTumor on four public datasets - BraTS, LiTS, KiTS, and MSD-Pancreas - covering diverse organs and imaging modalities. Experimental results demonstrate that our method outperforms state-of-the-art baselines, achieving superior image fidelity and segmentation accuracy. Our contributions include a novel integration of sketches with diffusion models for medical image editing, fine-grained control over tumor progression visualization, and extensive validation across multiple datasets, setting a new benchmark in the field.
在医学成像中准确可视化和编辑肿瘤进展对于诊断、治疗计划和临床沟通至关重要。为了解决现有方法的主观性和精度有限等挑战,我们提出了SkEditTumor,这是一种基于草图扩散的可控肿瘤进展编辑模型。通过利用草图作为结构先验,我们的方法能够在保持结构完整性和视觉真实感的同时,精确修改肿瘤区域。我们在四个公共数据集(包括覆盖多个器官和成像方式的BraTS、LiTS、KiTS和MSD-Pancreas)上评估了SkEditTumor。实验结果表明,我们的方法优于最新基线,在图像保真度和分割精度方面取得了优越性。我们的贡献包括将草图与扩散模型相结合进行医学图像编辑的新颖集成,对肿瘤进展可视化的精细控制,以及在多个数据集上的广泛验证,为这一领域树立了新的基准。
论文及项目相关链接
PDF 9 pages, 4 figures
Summary
本文提出一种基于草图扩散模型的肿瘤进展编辑方法SkEditTumor,通过利用草图作为结构先验,实现对肿瘤区域的精确修改,同时保持结构完整性和视觉真实性。在多个公共数据集上的实验结果表明,该方法在图像保真度和分割精度方面优于现有技术基线,为医学图像编辑领域树立了新的标杆。
Key Takeaways
- SkEditTumor是一种基于草图扩散模型的肿瘤进展编辑方法,可精确修改肿瘤区域。
- Sketches被用作结构先验,使方法能够在保持结构完整性和视觉真实性的同时,对肿瘤区域进行精确修改。
- SkEditTumor在多个公共数据集上进行了实验验证,包括BraTS、LiTS、KiTS和MSD-Pancreas,覆盖不同器官和成像模式。
- 与现有技术基线相比,SkEditTumor在图像保真度和分割精度方面表现出优越性。
- SkEditTumor为医学图像编辑领域树立了新的标杆,特别是在肿瘤进展可视化和编辑方面。
- SkEditTumor集成了草图与扩散模型,提供了一种新颖的医学图像编辑方式。
点此查看论文截图

New CCD Driving Technique to Suppress Anomalous Charge Intrusion from Outside the Imaging Area for Soft X-ray Imager of Xtend onboard XRISM
Authors:Hirofumi Noda, Mio Aoyagi, Koji Mori, Hiroshi Tomida, Hiroshi Nakajima, Takaaki Tanaka, Hiromasa Suzuki, Hiroshi Murakami, Hiroyuki Uchida, Takeshi G. Tsuru, Keitaro Miyazaki, Kohei Kusunoki, Yoshiaki Kanemaru, Yuma Aoki, Kumiko Nobukawa, Masayoshi Nobukawa, Kohei Shima, Marina Yoshimoto, Kazunori Asakura, Hironori Matsumoto, Tomokage Yoneyama, Shogo B. Kobayashi, Kouichi Hagino, Hideki Uchiyama, Kiyoshi Hayashida
The Soft X-ray Imager (SXI) is an X-ray CCD camera of the Xtend system onboard the X-Ray Imaging and Spectroscopy Mission (XRISM), which was successfully launched on September 7, 2023 (JST). During ground cooling tests of the CCDs in 2020/2021, using the flight-model detector housing, electronic boards, and a mechanical cooler, we encountered an unexpected issue. Anomalous charges appeared outside the imaging area of the CCDs and intruded into the imaging area, causing pulse heights to stick to the maximum value over a wide region. Although this issue has not occurred in subsequent tests or in orbit so far, it could seriously affect the imaging and spectroscopic performance of the SXI if it were to happen in the future. Through experiments with non-flight-model detector components, we successfully reproduced the issue and identified that the anomalous charges intrude via the potential structure created by the charge injection electrode at the top of the imaging area. To prevent anomalous charge intrusion and maintain imaging and spectroscopic performance that satisfies the requirements, even if this issue occurs in orbit, we developed a new CCD driving technique. This technique is different from the normal operation in terms of potential structure and its changes during imaging and charge injection. In this paper, we report an overview of the anomalous charge issue, the related potential structures, the development of the new CCD driving technique to prevent the issue, the imaging and spectroscopic performance of the new technique, and the results of experiments to investigate the cause of anomalous charges.
软X射线成像仪(SXI)是X射线成像与光谱任务(XRISM)上的X射线CCD相机,XRISM于2023年9月7日(日本标准时间)成功发射。在2020/2021年对CCD的地面冷却测试中,我们使用了飞行模型检测器外壳、电子板和机械冷却器,遇到了一个意料之外的问题。异常电荷出现在CCD成像区域之外,并侵入成像区域,导致脉冲高度在宽区域内达到最大值。虽然这一问题在随后的测试或在轨运行中尚未出现,但如果未来再次发生,可能会严重影响SXI的成像和光谱性能。我们通过使用非飞行模型检测器组件进行实验,成功复现了这一问题,并确定异常电荷是通过成像区域顶部的电荷注入电极产生的电位结构侵入的。为了预防异常电荷侵入并满足成像和光谱性能的要求,即使在未来在轨出现这一问题,我们也开发了一种新的CCD驱动技术。这种技术在电位结构和成像及电荷注入期间的变化方面与正常操作不同。本文概述了异常电荷问题、相关的电位结构、为防止该问题而开发的新CCD驱动技术、新技术的成像和光谱性能以及调查异常电荷原因的试验结果。
论文及项目相关链接
PDF 13 pages, 8 figures, Accepted for publication in JATIS XRISM special issue
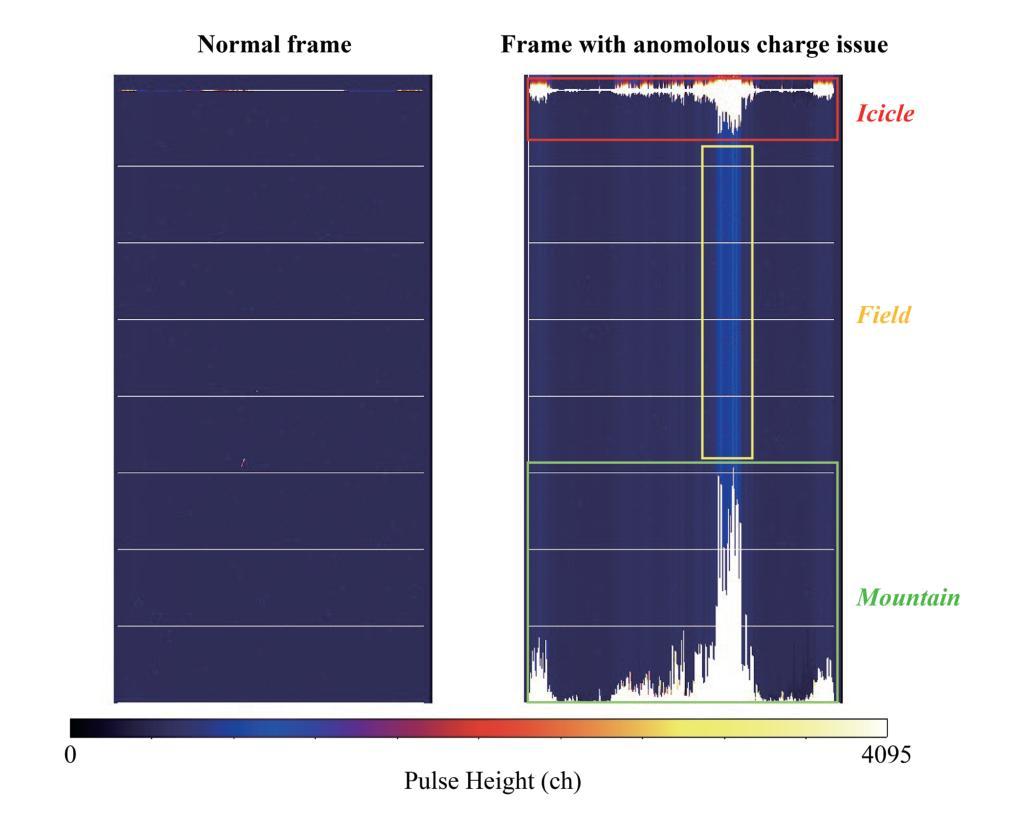
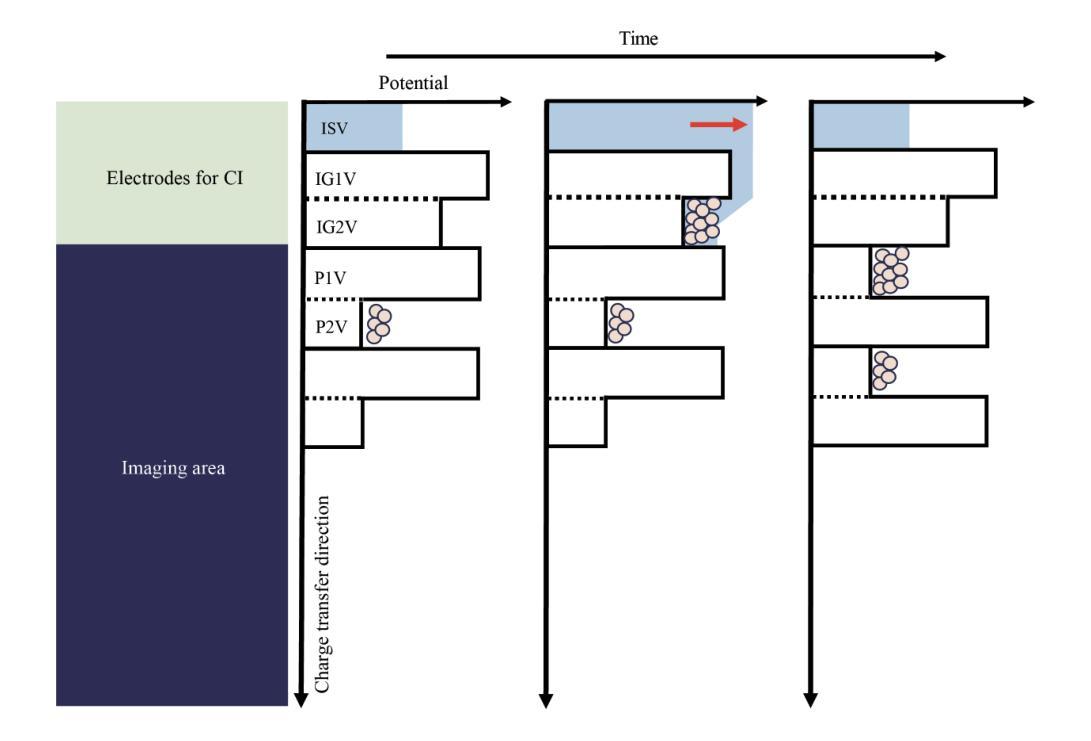
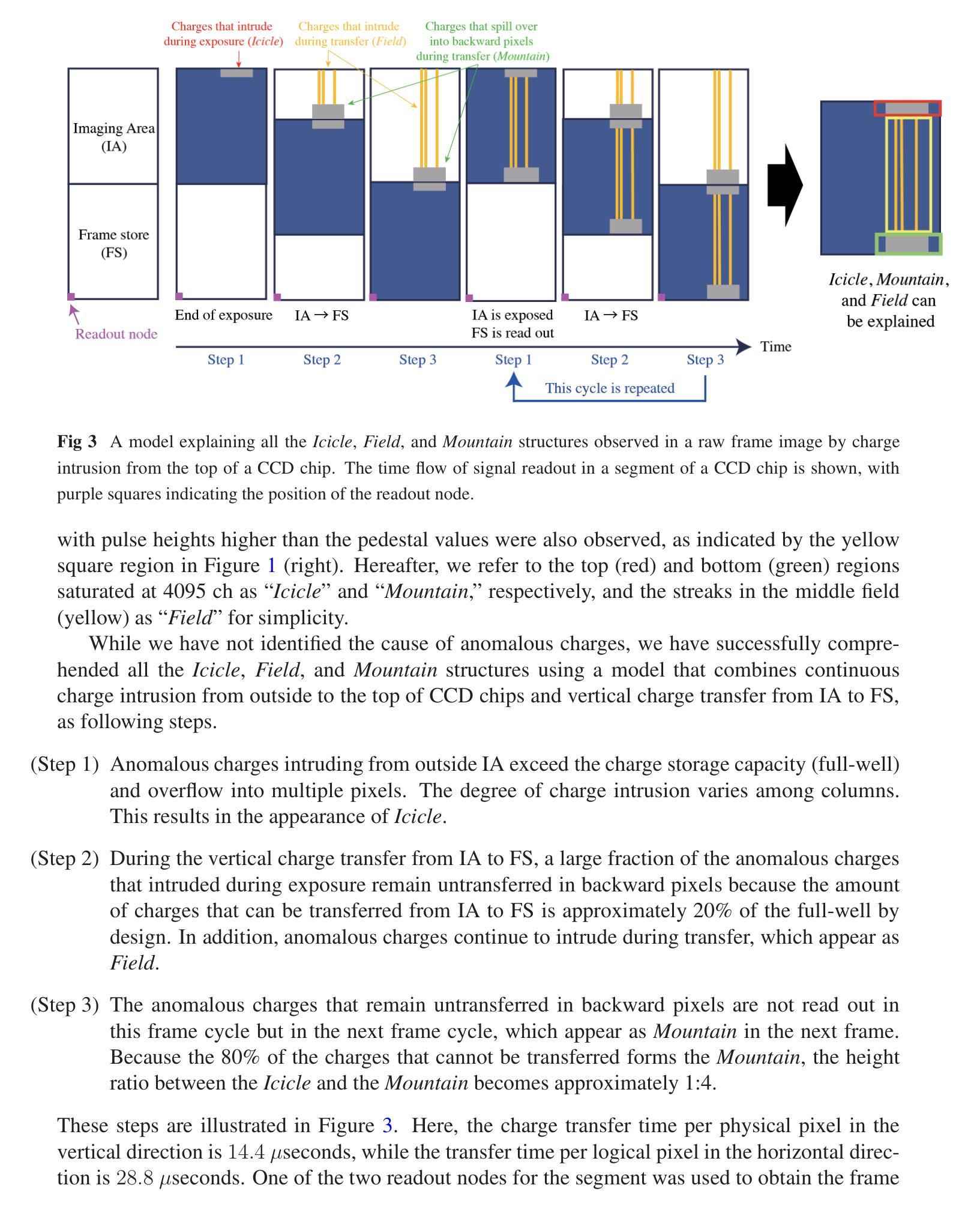
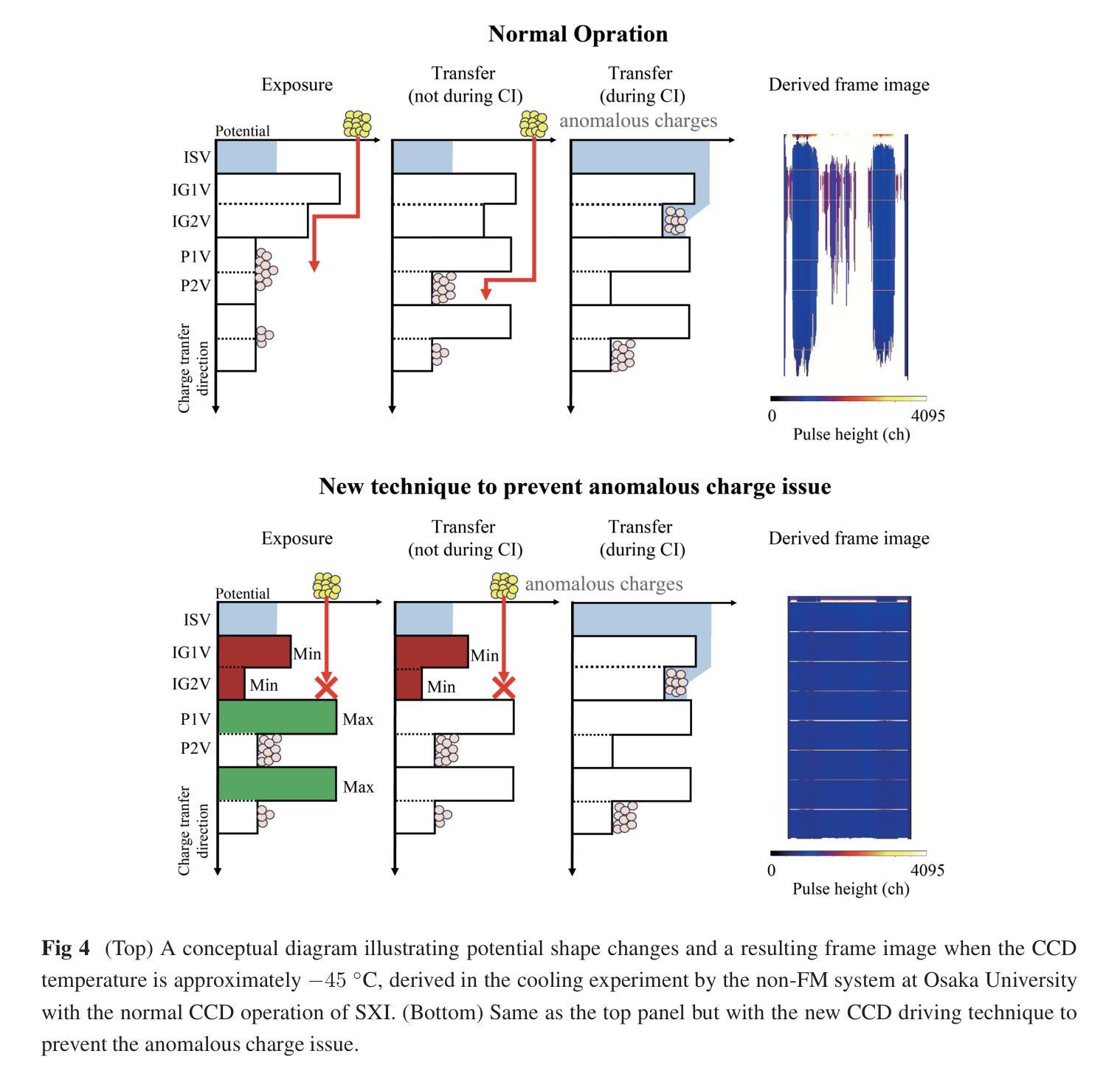
摘要
SXI是X射线成像与光谱任务(XRISM)的Xtend系统搭载的一款X射线CCD相机,于日本标准时间(JST)2023年9月7日成功发射。在地面冷却测试中,我们发现CCD成像区域外的异常电荷侵入成像区域,导致脉冲高度大面积达到最大值。尽管后续测试和轨道运行均未出现此问题,但若未来发生将严重影响成像和光谱性能。我们成功重现了这一问题并发现异常电荷通过成像区域顶部的电荷注入电极产生的潜在结构侵入。为解决此问题并保持成像和光谱性能满足要求,我们开发了一种新型CCD驱动技术。该技术通过改变成像和电荷注入期间的潜在结构和变化来防止异常电荷侵入。本文报告了异常电荷问题概述、相关潜在结构、防止问题的新型CCD技术开发、新技术的成像和光谱性能以及调查异常电荷原因的实验结果。
关键见解
- SXI是XRISM任务的X射线CCD相机,于2023年成功发射。
- 在地面冷却测试中发现了异常电荷侵入的问题,导致成像区域受到干扰。
- 异常电荷通过电荷注入电极产生的潜在结构侵入成像区域。
- 开发了一种新型CCD驱动技术,以预防异常电荷侵入并维持成像和光谱性能。
- 新技术改变了成像和电荷注入期间的潜在结构和变化。
- 该技术成功解决了异常电荷侵入的问题,提高了成像质量。
- 文中还报告了调查异常电荷原因的实验结果。
点此查看论文截图

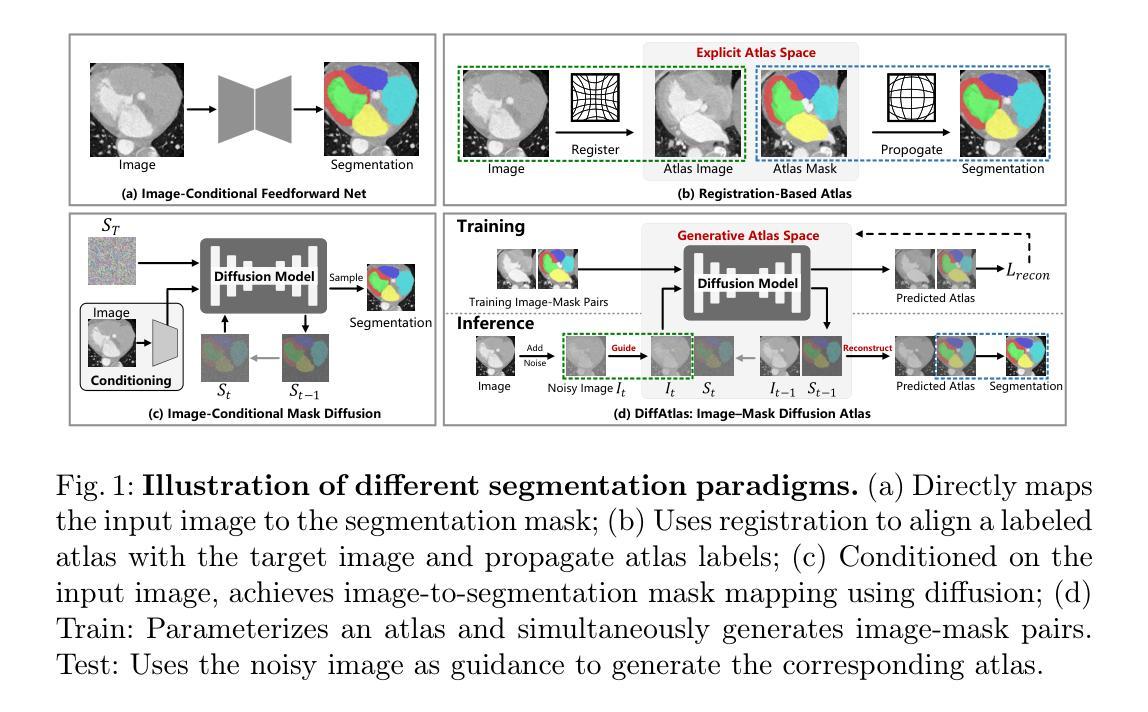
DiffAtlas: GenAI-fying Atlas Segmentation via Image-Mask Diffusion
Authors:Hantao Zhang, Yuhe Liu, Jiancheng Yang, Weidong Guo, Xinyuan Wang, Pascal Fua
Accurate medical image segmentation is crucial for precise anatomical delineation. Deep learning models like U-Net have shown great success but depend heavily on large datasets and struggle with domain shifts, complex structures, and limited training samples. Recent studies have explored diffusion models for segmentation by iteratively refining masks. However, these methods still retain the conventional image-to-mask mapping, making them highly sensitive to input data, which hampers stability and generalization. In contrast, we introduce DiffAtlas, a novel generative framework that models both images and masks through diffusion during training, effectively ``GenAI-fying’’ atlas-based segmentation. During testing, the model is guided to generate a specific target image-mask pair, from which the corresponding mask is obtained. DiffAtlas retains the robustness of the atlas paradigm while overcoming its scalability and domain-specific limitations. Extensive experiments on CT and MRI across same-domain, cross-modality, varying-domain, and different data-scale settings using the MMWHS and TotalSegmentator datasets demonstrate that our approach outperforms existing methods, particularly in limited-data and zero-shot modality segmentation. Code is available at https://github.com/M3DV/DiffAtlas.
精确医学图像分割对于精确解剖界定至关重要。U-Net等深度学习模型已经取得了巨大的成功,但它们严重依赖于大型数据集,在领域转移、复杂结构和有限训练样本方面遇到了困难。最近的研究探索了通过迭代细化掩膜进行分割的扩散模型。然而,这些方法仍然保留了传统的图像到掩膜映射,使它们对输入数据高度敏感,这阻碍了稳定性和泛化能力。相比之下,我们引入了DiffAtlas,这是一种新型生成框架,在训练过程中通过扩散对图像和掩膜进行建模,有效地实现了基于图谱的分割的“GenAI化”。在测试过程中,该模型被引导生成特定的目标图像-掩膜对,从中获得相应的掩膜。DiffAtlas保留了图谱范式的稳健性,同时克服了其可扩展性和领域特定限制。在MMWHS和TotalSegmentator数据集上进行的关于CT和MRI的同域、跨模态、变域和不同数据规模设置的广泛实验表明,我们的方法优于现有方法,特别是在有限数据和零样本模态分割方面。代码可在https://github.com/M3DV/DiffAtlas获得。
论文及项目相关链接
PDF 11 pages
Summary
本文介绍了DiffAtlas,一种新型的医学图像分割生成框架。该框架在训练过程中通过扩散建模图像和掩膜,有效实现了基于图谱的分割“GenAI化”。通过实验验证,DiffAtlas在相同领域、跨模态、不同领域和不同数据规模设置下的CT和MRI图像分割中表现出卓越性能,特别是在有限数据和零样本模态分割方面。
Key Takeaways
- 医学图像分割的准确性对于精确解剖结构划分至关重要。
- U-Net等深度学习模型虽然表现出色,但在处理领域变化、复杂结构和有限训练样本时存在挑战。
- 扩散模型在图像分割中的应用通过迭代优化掩膜来改进分割效果。
- 现有方法仍然依赖于传统的图像到掩膜映射,对输入数据敏感,影响稳定性和泛化能力。
- DiffAtlas框架通过建模图像和掩膜的扩散过程,实现了基于图谱的分割“GenAI化”。
- DiffAtlas在多种设置下的实验表现优异,特别是在有限数据和零样本模态分割方面。
点此查看论文截图

Continuous Online Adaptation Driven by User Interaction for Medical Image Segmentation
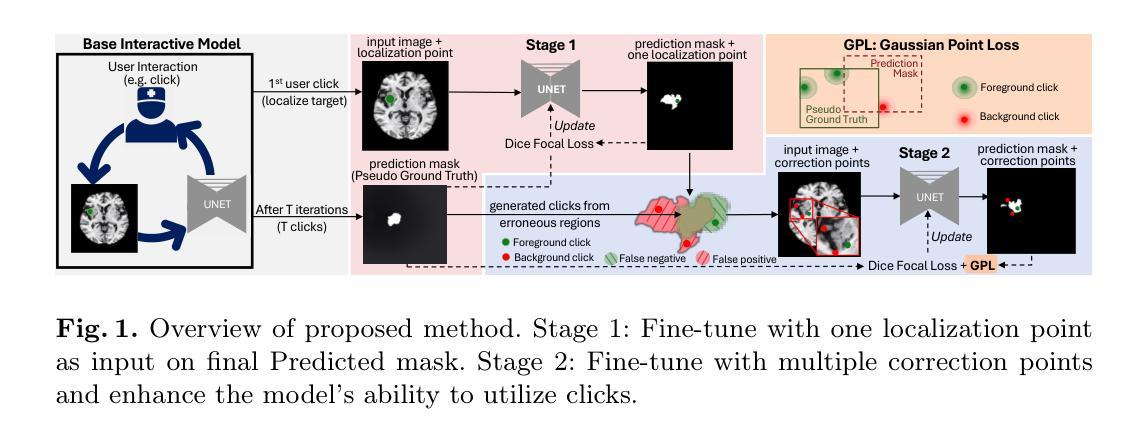
Authors:Wentian Xu, Ziyun Liang, Harry Anthony, Yasin Ibrahim, Felix Cohen, Guang Yang, Daniel Whitehouse, David Menon, Virginia Newcombe, Konstantinos Kamnitsas
Interactive segmentation models use real-time user interactions, such as mouse clicks, as extra inputs to dynamically refine the model predictions. After model deployment, user corrections of model predictions could be used to adapt the model to the post-deployment data distribution, countering distribution-shift and enhancing reliability. Motivated by this, we introduce an online adaptation framework that enables an interactive segmentation model to continuously learn from user interaction and improve its performance on new data distributions, as it processes a sequence of test images. We introduce the Gaussian Point Loss function to train the model how to leverage user clicks, along with a two-stage online optimization method that adapts the model using the corrected predictions generated via user interactions. We demonstrate that this simple and therefore practical approach is very effective. Experiments on 5 fundus and 4 brain MRI databases demonstrate that our method outperforms existing approaches under various data distribution shifts, including segmentation of image modalities and pathologies not seen during training.
交互式分割模型使用实时用户交互,如鼠标点击,作为额外的输入来动态优化模型预测。在模型部署后,用户可以对模型预测进行修正,使模型适应部署后的数据分布,对抗分布偏移,提高可靠性。受此启发,我们引入了一个在线适配框架,使交互式分割模型能够在处理测试图像序列时,从用户交互中持续学习,并提高其在新的数据分布上的性能。我们引入了高斯点损失函数来训练模型如何利用用户点击,以及一个两阶段的在线优化方法,该方法使用用户交互生成的修正预测来适应模型。我们证明这种简单实用的方法非常有效。在5个眼底和4个脑部MRI数据库上的实验表明,我们的方法在多种数据分布偏移情况下,包括图像模态和训练期间未见到的病理分割,都优于现有方法。
论文及项目相关链接
Summary
实时交互分割模型通过利用用户实时交互(如鼠标点击)作为额外输入,动态优化模型预测。部署模型后,用户修正的模型预测可用于适应部署后的数据分布,对抗分布偏移,提高可靠性。我们引入了一个在线适应框架,使交互分割模型能够从用户交互中学习,并在处理测试图像序列时,适应新数据分布并改进性能。通过高斯点损失函数训练模型利用用户点击,并提出两阶段在线优化方法,使用用户交互生成的修正预测来适应模型。在5个眼底和4个脑部MRI数据库上的实验表明,该方法在各种数据分布偏移下的性能优于现有方法,包括图像模态和病理的分割,这些在训练时并未见过。
Key Takeaways
- 实时交互分割模型利用用户实时交互作为额外输入来动态优化模型预测。
- 用户修正的模型预测可以用于适应模型到部署后的数据分布。
- 引入了一个在线适应框架,使模型能够从用户交互中学习并改进性能。
- 高斯点损失函数被用来训练模型如何有效利用用户点击。
- 提出了一种两阶段的在线优化方法,通过用户交互生成的修正预测来适应模型。
- 该方法在多种数据分布偏移情况下的性能优于现有方法。
点此查看论文截图

Dynamic Dictionary Learning for Remote Sensing Image Segmentation
Authors:Xuechao Zou, Yue Li, Shun Zhang, Kai Li, Shiying Wang, Pin Tao, Junliang Xing, Congyan Lang
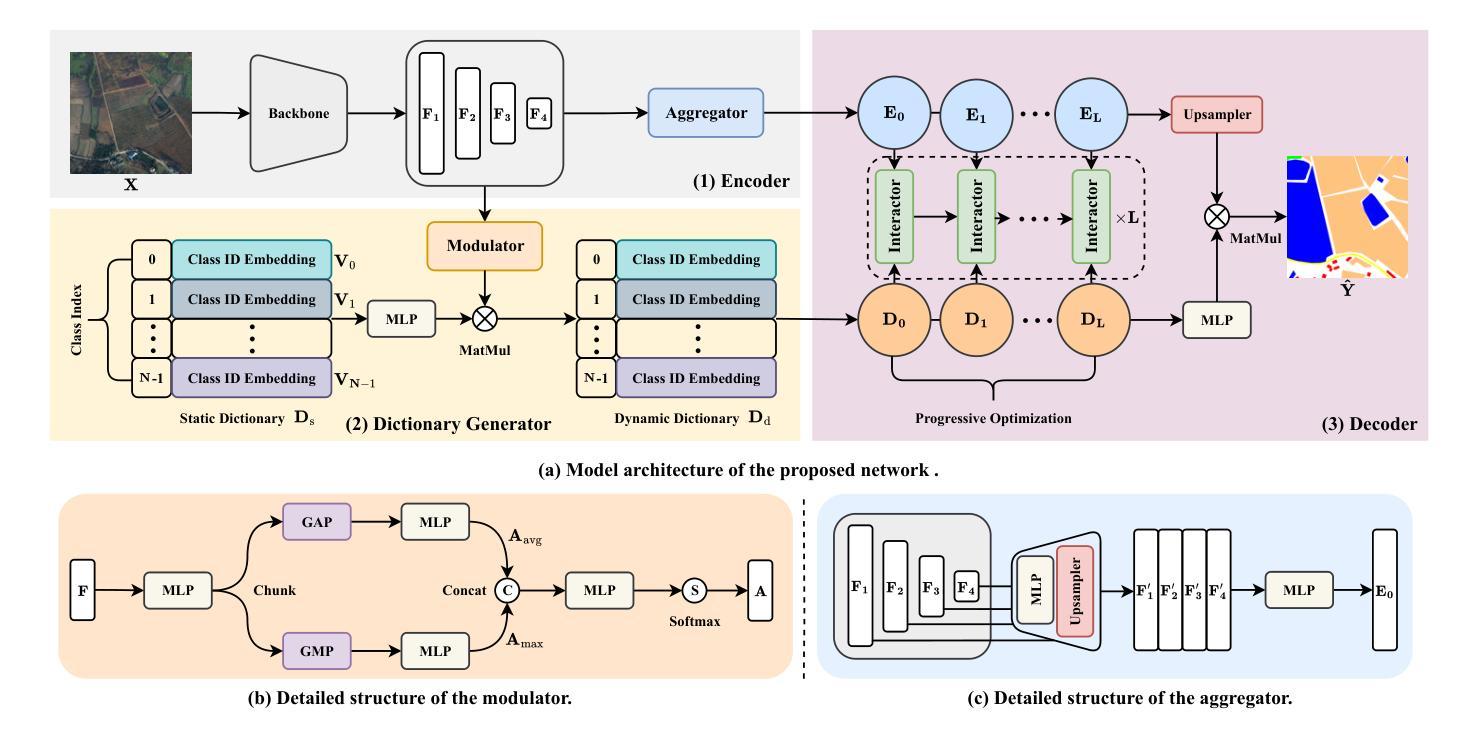
Remote sensing image segmentation faces persistent challenges in distinguishing morphologically similar categories and adapting to diverse scene variations. While existing methods rely on implicit representation learning paradigms, they often fail to dynamically adjust semantic embeddings according to contextual cues, leading to suboptimal performance in fine-grained scenarios such as cloud thickness differentiation. This work introduces a dynamic dictionary learning framework that explicitly models class ID embeddings through iterative refinement. The core contribution lies in a novel dictionary construction mechanism, where class-aware semantic embeddings are progressively updated via multi-stage alternating cross-attention querying between image features and dictionary embeddings. This process enables adaptive representation learning tailored to input-specific characteristics, effectively resolving ambiguities in intra-class heterogeneity and inter-class homogeneity. To further enhance discriminability, a contrastive constraint is applied to the dictionary space, ensuring compact intra-class distributions while maximizing inter-class separability. Extensive experiments across both coarse- and fine-grained datasets demonstrate consistent improvements over state-of-the-art methods, particularly in two online test benchmarks (LoveDA and UAVid). Code is available at https://anonymous.4open.science/r/D2LS-8267/.
遥感图像分割在区分形态相似类别和适应多种场景变化方面持续面临挑战。尽管现有方法依赖于隐式表示学习范式,但它们通常无法根据上下文线索动态调整语义嵌入,导致在云厚度差异等精细场景中的性能不佳。本文引入了一种动态字典学习框架,通过迭代细化显式建模类ID嵌入。核心贡献在于一种新的字典构建机制,其中通过图像特征和字典嵌入之间的多阶段交替交叉注意力查询,逐步更新类感知语义嵌入。这一过程使适应输入特定特性的自适应表示学习成为可能,有效地解决了类内异质性和类间同质性中的歧义。为了进一步提高鉴别力,在字典空间应用对比约束,确保类内分布紧凑,同时最大化类间可分性。在粗细粒度数据集上的大量实验表明,与最新方法相比,特别是在两个在线测试基准(LoveDA和UAVid)上,改进具有一致性。代码可在https://anonymous.4open.science/r/D2LS-8267/找到。
论文及项目相关链接
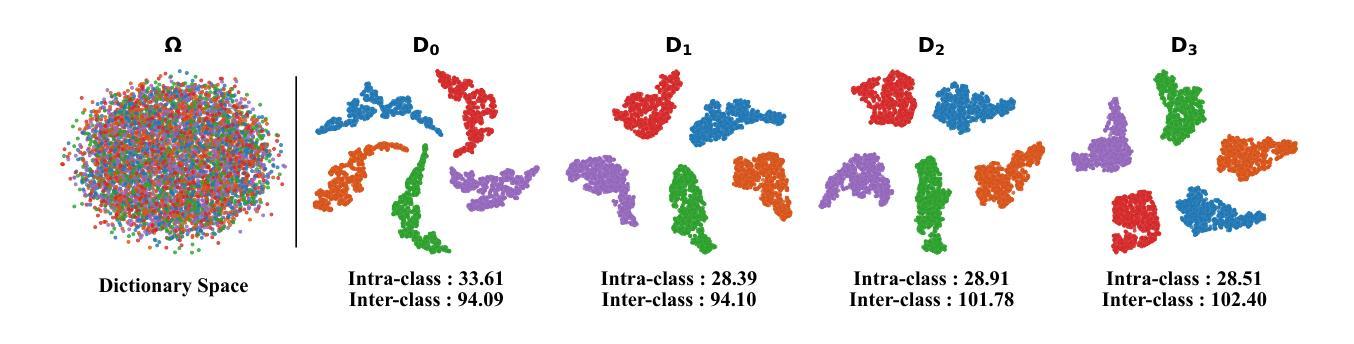
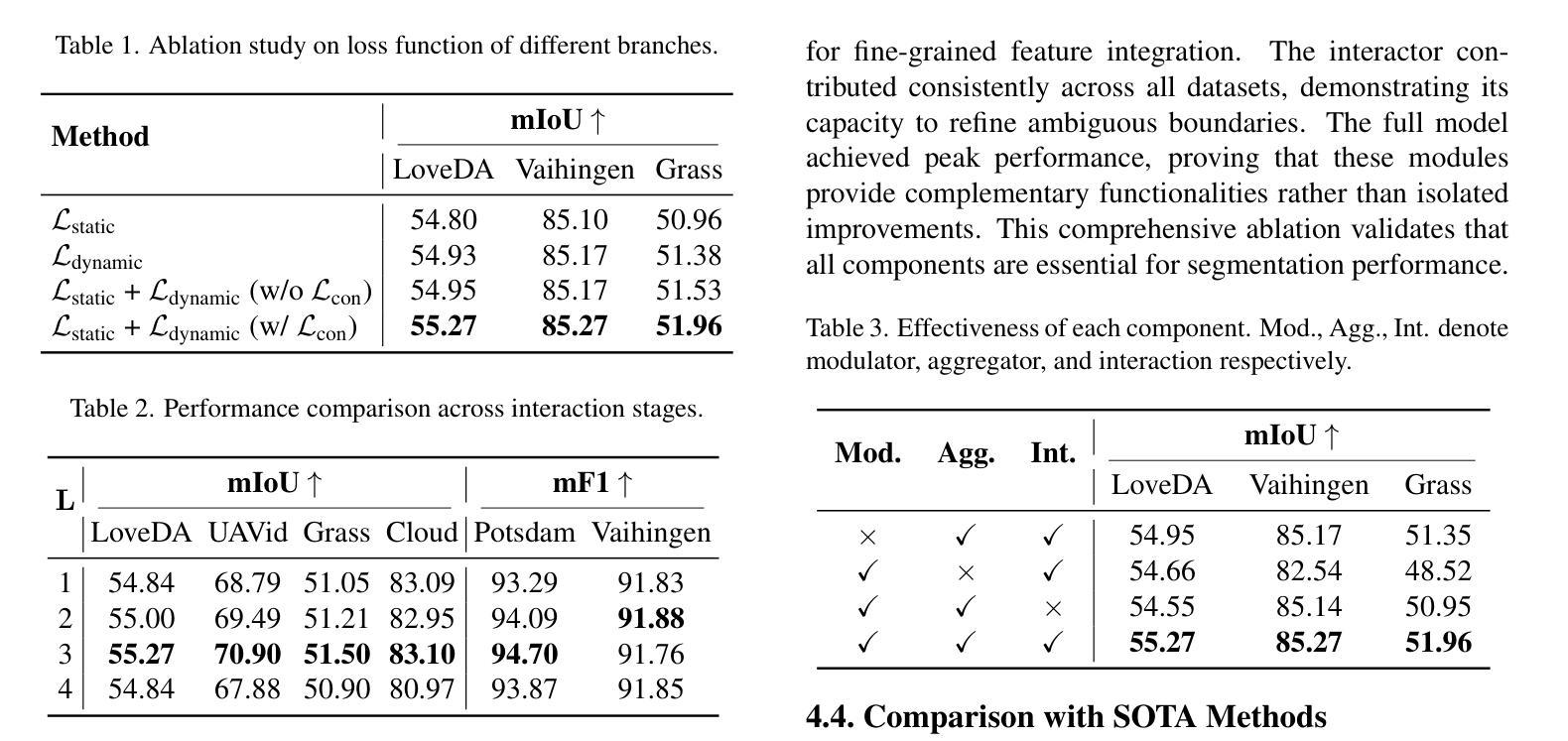
Summary
本文提出一种动态字典学习框架,通过迭代优化显式建模类别ID嵌入,解决遥感图像分割中形态相似类别区分及场景多样变化适应问题。框架核心在于新颖字典构建机制,通过图像特征与字典嵌入之间的多阶段交替跨注意力查询,逐步更新类别感知语义嵌入,实现自适应表示学习,有效解冑类内异质性和类间同质性引起的歧义。为提升鉴别力,应用对比约束于字典空间,确保类内分布紧凑并最大化类间可分性。在粗细粒度数据集上的广泛实验表明,该方法较前沿方法有一致性改进,特别是在两个在线测试基准(LoveDA和UAVid)上表现优异。
Key Takeaways
- 遥感图像分割面临区分形态相似类别和适应场景多样性的挑战。
- 现有方法依赖隐式表示学习,无法根据上下文动态调整语义嵌入。
- 本文提出动态字典学习框架,通过迭代优化显式建模类别ID嵌入。
- 框架核心在于新颖的字典构建机制,逐步更新类别感知语义嵌入。
- 实现自适应表示学习,有效解决类内异质性和类间同质性问题。
- 通过应用对比约束于字典空间,提升鉴别力。
点此查看论文截图

LSA: Latent Style Augmentation Towards Stain-Agnostic Cervical Cancer Screening
Authors:Jiangdong Cai, Haotian Jiang, Zhenrong Shen, Yonghao Li, Honglin Xiong, Lichi Zhang, Qian Wang
The deployment of computer-aided diagnosis systems for cervical cancer screening using whole slide images (WSIs) faces critical challenges due to domain shifts caused by staining variations across different scanners and imaging environments. While existing stain augmentation methods improve patch-level robustness, they fail to scale to WSIs due to two key limitations: (1) inconsistent stain patterns when extending patch operations to gigapixel slides, and (2) prohibitive computational/storage costs from offline processing of augmented WSIs.To address this, we propose Latent Style Augmentation (LSA), a framework that performs efficient, online stain augmentation directly on WSI-level latent features. We first introduce WSAug, a WSI-level stain augmentation method ensuring consistent stain across patches within a WSI. Using offline-augmented WSIs by WSAug, we design and train Stain Transformer, which can simulate targeted style in the latent space, efficiently enhancing the robustness of the WSI-level classifier. We validate our method on a multi-scanner WSI dataset for cervical cancer diagnosis. Despite being trained on data from a single scanner, our approach achieves significant performance improvements on out-of-distribution data from other scanners. Code will be available at https://github.com/caijd2000/LSA.
利用全切片图像(WSI)进行宫颈癌筛查的计算机辅助诊断系统的部署,由于不同扫描仪和成像环境中的染色变化导致的领域漂移而面临重大挑战。现有的染色增强方法虽然提高了补丁级别的稳健性,但由于两个关键局限性而无法扩展到全切片图像:一是将补丁操作扩展到千兆像素切片时,染色模式不一致;二是通过离线处理增强全切片图像的计算和存储成本过高。为了解决这一问题,我们提出了潜在风格增强(LSA)框架,该框架在全切片图像级别的潜在特征上直接进行高效在线染色增强。我们首先引入了WSAug全切片图像级别的染色增强方法,确保全切片图像内各补丁之间的染色一致性。通过WSAug对离线增强的全切片图像进行处理,我们设计和训练了染色转换器,可以在潜在空间中模拟目标风格,有效提高全切片图像级别分类器的稳健性。我们在多扫描仪全切片图像数据集上验证了我们的方法,用于宫颈癌诊断。尽管我们的方法是在单一扫描仪的数据上进行训练的,但在其他扫描仪的离分布数据上取得了显著的性能提升。代码将在https://github.com/caijd2000/LSA上提供。
论文及项目相关链接
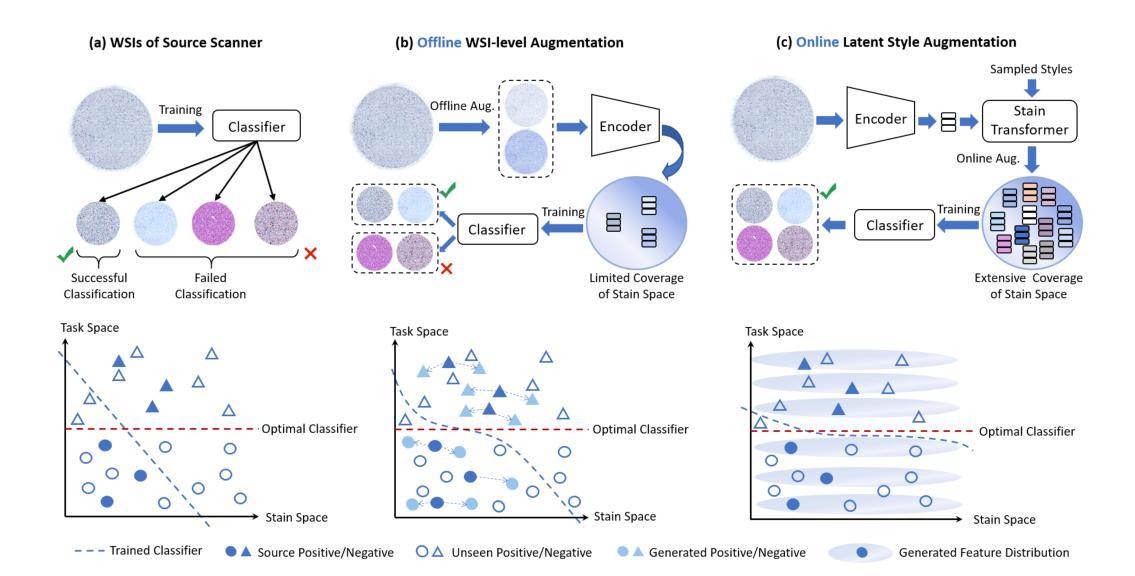
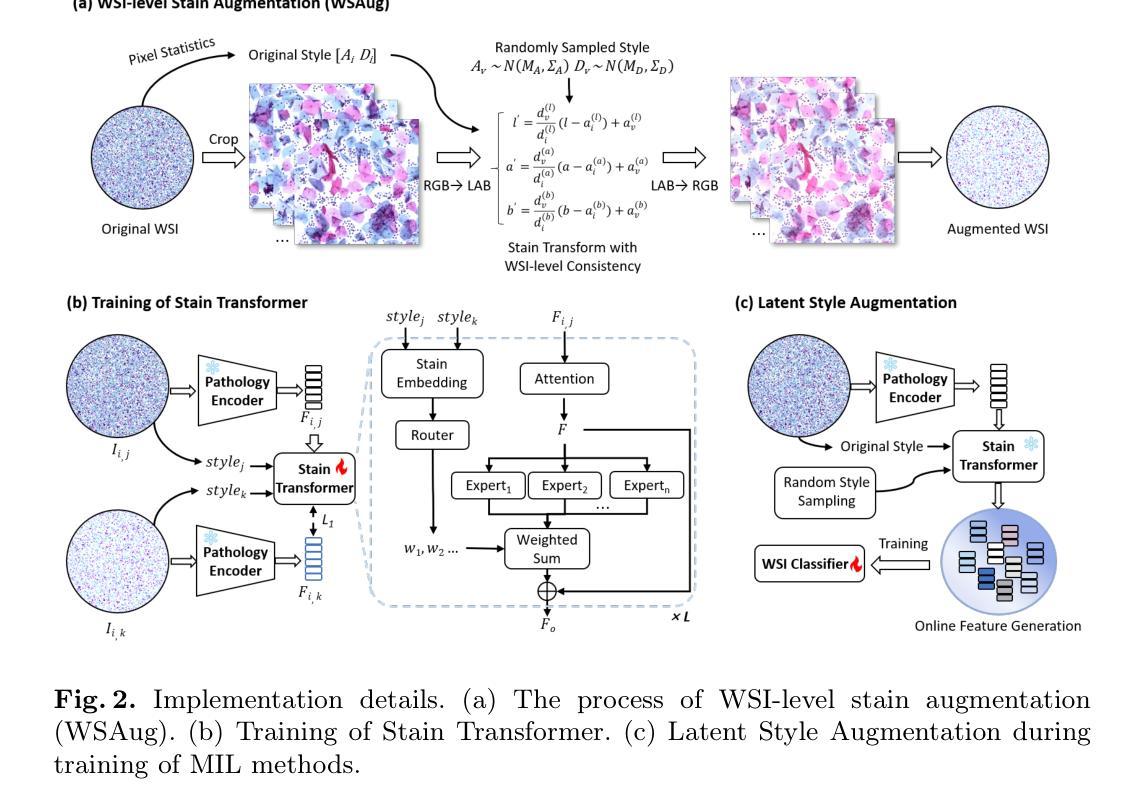
Summary
计算机辅助诊断系统在宫颈癌筛查中使用全切片图像(WSI)时面临领域偏移的挑战,因为不同扫描仪和成像环境的染色变化导致的。现有的染色增强方法在补丁级别上提高了鲁棒性,但由于关键局限无法扩展到WSI。我们提出潜态染色增强(LSA)框架,直接在WSI级别的潜在特征上进行高效的在线染色增强。通过引入WSAug染色增强方法和Stain Transformer,我们设计了一个能够在潜在空间中模拟目标风格的模型,有效提高了WSI级别分类器的稳健性。在多个扫描仪的WSI数据集上进行验证,我们的方法在来自其他扫描仪的离群数据上取得了显著的性能提升。
Key Takeaways
- 计算机辅助诊断系统在使用全切片图像进行宫颈癌筛查时面临染色变化导致的领域偏移挑战。
- 现有染色增强方法无法扩展到全切片图像,存在补丁级别的不一致性和高计算存储成本问题。
- 潜态染色增强(LSA)框架提出在线染色增强方法,直接在WSI级别上进行。
- WSAug染色增强方法确保全切片图像内补丁间染色的一致性。
- Stain Transformer设计用于模拟潜在空间中的目标风格,提高WSI级别分类器的稳健性。
- 在多扫描仪WSI数据集上的验证显示,该方法在来自其他扫描仪的离群数据上取得了显著性能提升。
点此查看论文截图

PathVQ: Reforming Computational Pathology Foundation Model for Whole Slide Image Analysis via Vector Quantization
Authors:Honglin Li, Zhongyi Shui, Yunlong Zhang, Chenglu Zhu, Lin Yang
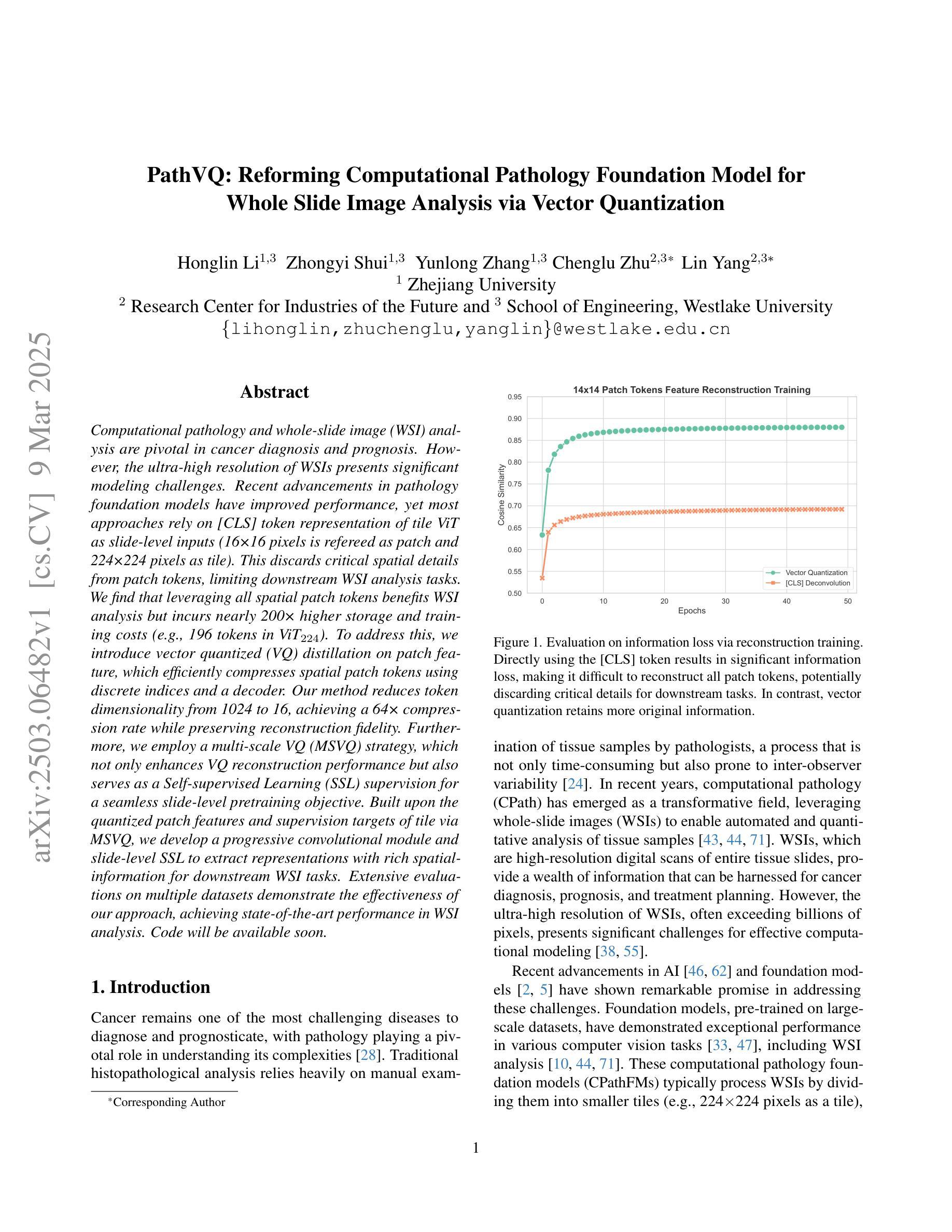
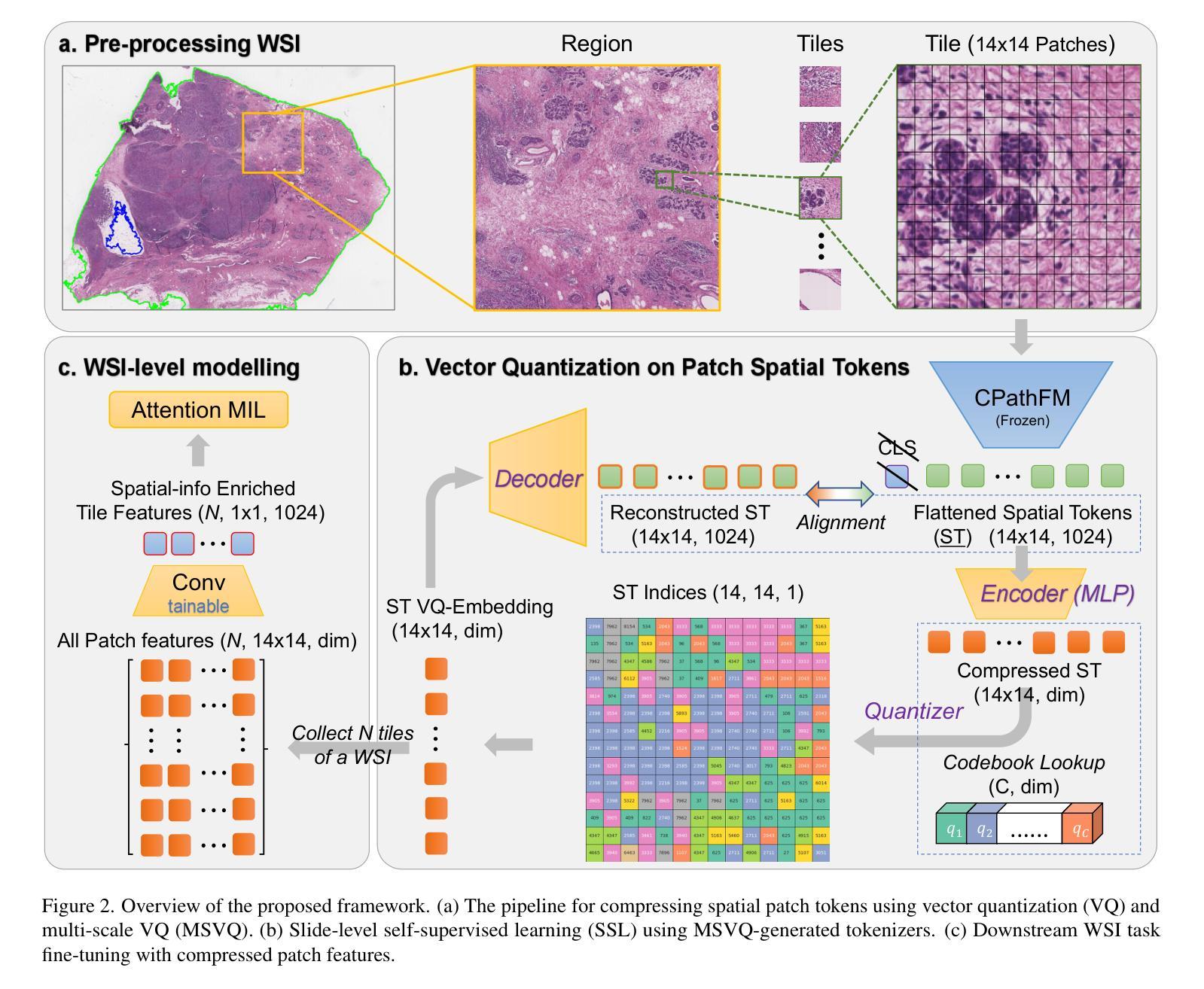
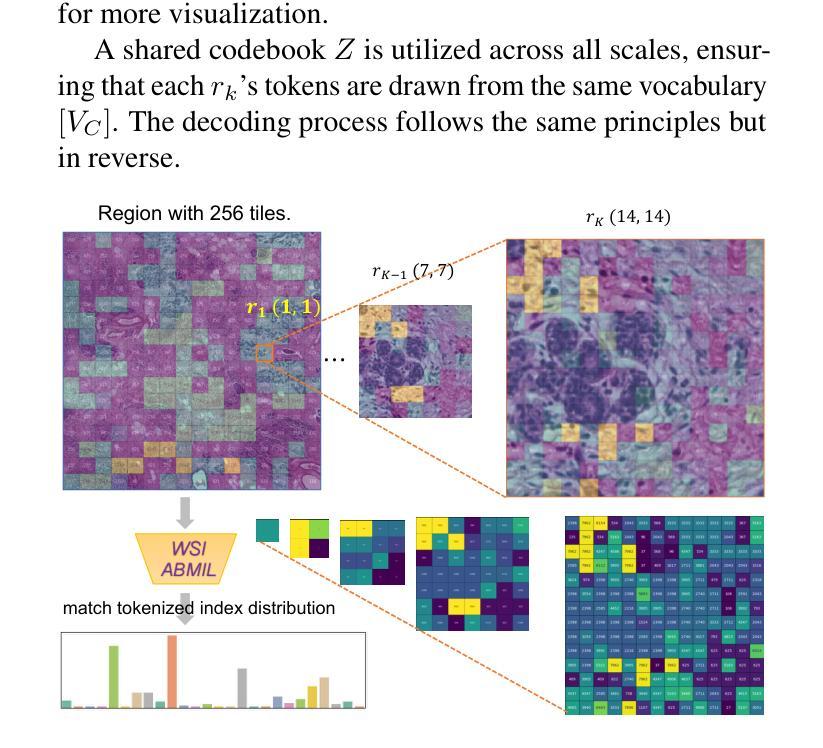
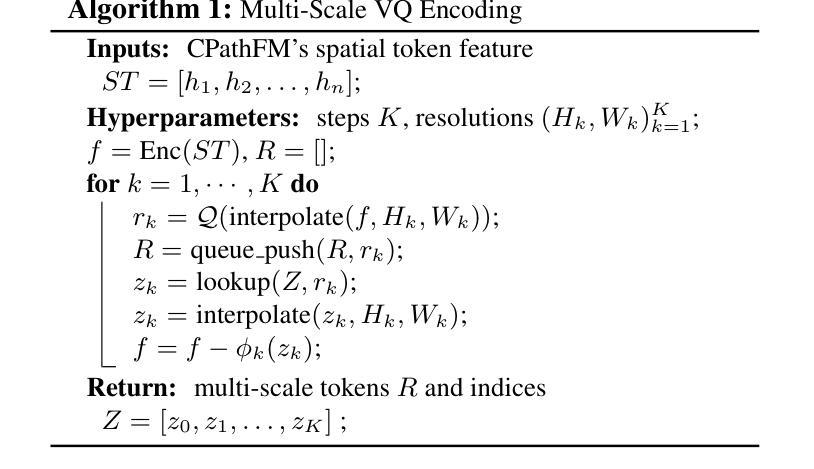
Computational pathology and whole-slide image (WSI) analysis are pivotal in cancer diagnosis and prognosis. However, the ultra-high resolution of WSIs presents significant modeling challenges. Recent advancements in pathology foundation models have improved performance, yet most approaches rely on [CLS] token representation of tile ViT as slide-level inputs (16x16 pixels is refereed as patch and 224x224 pixels as tile). This discards critical spatial details from patch tokens, limiting downstream WSI analysis tasks. We find that leveraging all spatial patch tokens benefits WSI analysis but incurs nearly 200x higher storage and training costs (e.g., 196 tokens in ViT$_{224}$). To address this, we introduce vector quantized (VQ) distillation on patch feature, which efficiently compresses spatial patch tokens using discrete indices and a decoder. Our method reduces token dimensionality from 1024 to 16, achieving a 64x compression rate while preserving reconstruction fidelity. Furthermore, we employ a multi-scale VQ (MSVQ) strategy, which not only enhances VQ reconstruction performance but also serves as a Self-supervised Learning (SSL) supervision for a seamless slide-level pretraining objective. Built upon the quantized patch features and supervision targets of tile via MSVQ, we develop a progressive convolutional module and slide-level SSL to extract representations with rich spatial-information for downstream WSI tasks. Extensive evaluations on multiple datasets demonstrate the effectiveness of our approach, achieving state-of-the-art performance in WSI analysis. Code will be available soon.
计算病理学和全切片图像(WSI)分析在癌症诊断和治疗中起着至关重要的作用。然而,WSI的超高分辨率给建模带来了巨大的挑战。尽管病理学基础模型的最新进展提高了性能,但大多数方法仍然依赖于将瓷砖ViT的[CLS]令牌表示作为切片级别的输入(其中,将16x16像素称为补丁,而224x224像素称为瓷砖)。这种做法丢弃了补丁令牌中的关键空间细节,限制了后续的WSI分析任务。我们发现利用所有空间补丁令牌对WSI分析是有益的,但这会导致存储和训练成本增加近200倍(例如,ViT中使用的令牌高达196个)。为了解决这一问题,我们引入了基于补丁特征的向量量化(VQ)蒸馏技术。使用离散索引和解码器,我们的方法可以高效地压缩空间补丁令牌,将令牌维度从1024降低到16,实现了高达64倍的压缩率,同时保持了重建保真度。此外,我们还采用了多尺度VQ(MSVQ)策略,这不仅提高了VQ重建性能,而且还作为一种自我监督学习(SSL)的监督手段,实现了无缝的切片级预训练目标。基于量化补丁特征和通过MSVQ获得的瓷砖监督目标,我们开发了一个渐进式卷积模块和切片级SSL,以提取具有丰富空间信息的表示用于下游WSI任务。在多个数据集上的广泛评估表明,我们的方法取得了最先进的效果。代码将在近期发布。
论文及项目相关链接
摘要
计算病理学和全幻灯片图像(WSI)分析在癌症诊断和治疗中至关重要。然而,WSI的超高分辨率带来了显著的建模挑战。虽然病理学基础模型的最新进展提高了性能,但大多数方法仍然依赖于[CLS]标记表示瓷砖ViT作为幻灯片级别的输入(以将一个幻灯片分割成若干块的方式),这种方式忽略了补丁令牌的空间细节,限制了下游WSI分析任务的效果。本研究通过利用所有空间补丁令牌对WSI分析进行改进,但同时也带来了近200倍的存储和训练成本增加。为解决此问题,我们引入了矢量量化(VQ)蒸馏补丁特征技术,该技术使用离散索引和解码器有效地压缩空间补丁令牌。我们的方法将令牌维度从1024减少到16,实现了64倍的压缩率,同时保持了重建保真度。此外,我们还采用了多尺度VQ(MSVQ)策略,不仅提高了VQ重建性能,还作为无缝幻灯片级别的自监督学习(SSL)监督,用于预训练目标。基于量化补丁特征和通过MSVQ获得的瓷砖监督目标,我们开发了一个渐进的卷积模块和幻灯片级别的SSL,以提取具有丰富空间信息的表示,用于下游WSI任务。在多个数据集上的广泛评估表明,我们的方法实现了WSI分析的最先进性能。代码将很快可用。
关键见解
- 计算病理学及全幻灯片图像分析在癌症诊断与预后评估中起关键作用。
- WSI的超高分辨率带来了建模挑战。
- 大多数现有的方法依赖于特定的标记表示方式(如ViT模型中的瓷砖级别输入),这限制了空间细节的捕获和分析效果。
- 直接利用所有空间细节(补丁令牌)虽然能改善分析效果,但成本显著增加。
- 提出矢量量化(VQ)蒸馏补丁特征技术,能在压缩空间补丁令牌的同时保持重建的保真度。这种技术显著减少了存储和训练成本。
- 多尺度VQ(MSVQ)策略不仅增强了VQ重建性能,还提供了自监督学习的预训练目标。
点此查看论文截图