⚠️ 以下所有内容总结都来自于 大语言模型的能力,如有错误,仅供参考,谨慎使用
🔴 请注意:千万不要用于严肃的学术场景,只能用于论文阅读前的初筛!
💗 如果您觉得我们的项目对您有帮助 ChatPaperFree ,还请您给我们一些鼓励!⭐️ HuggingFace免费体验
2025-04-19 更新
Multi-Parameter Molecular MRI Quantification using Physics-Informed Self-Supervised Learning
Authors:Alex Finkelstein, Nikita Vladimirov, Moritz Zaiss, Or Perlman
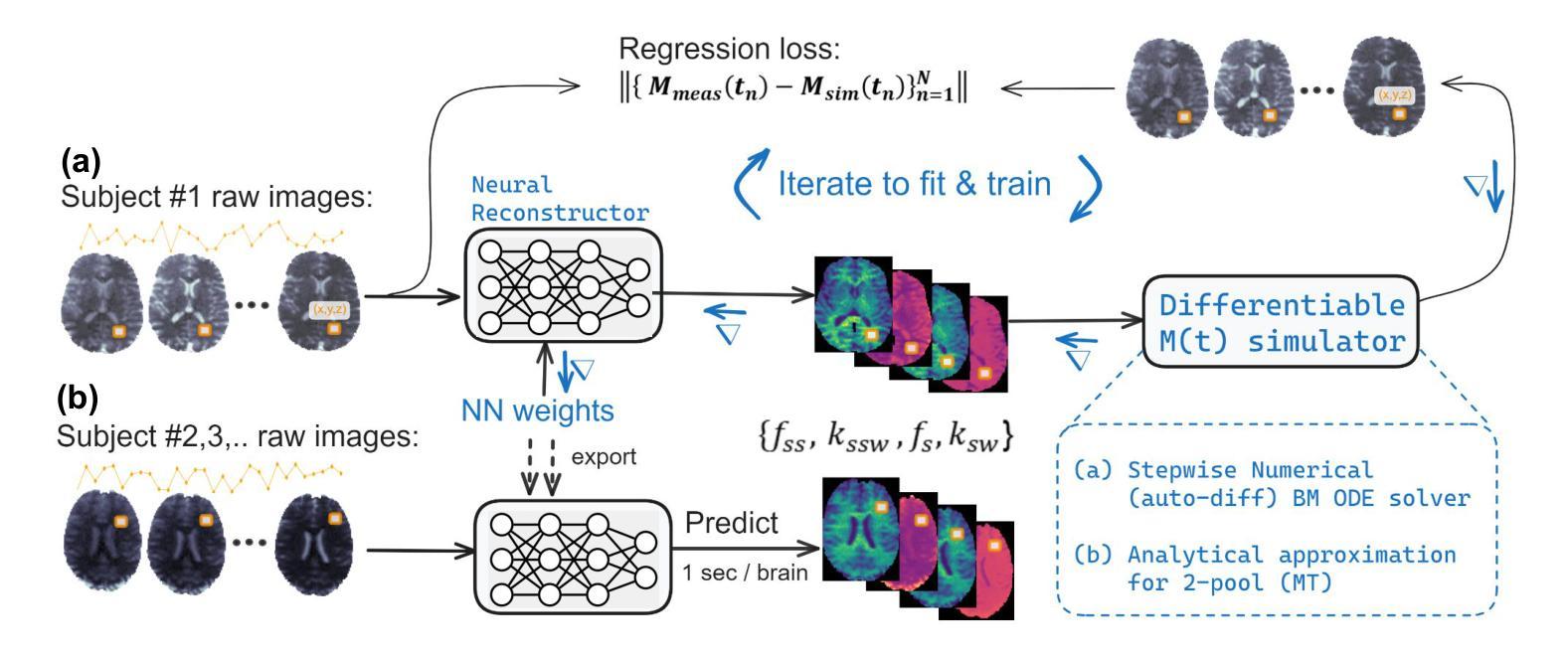
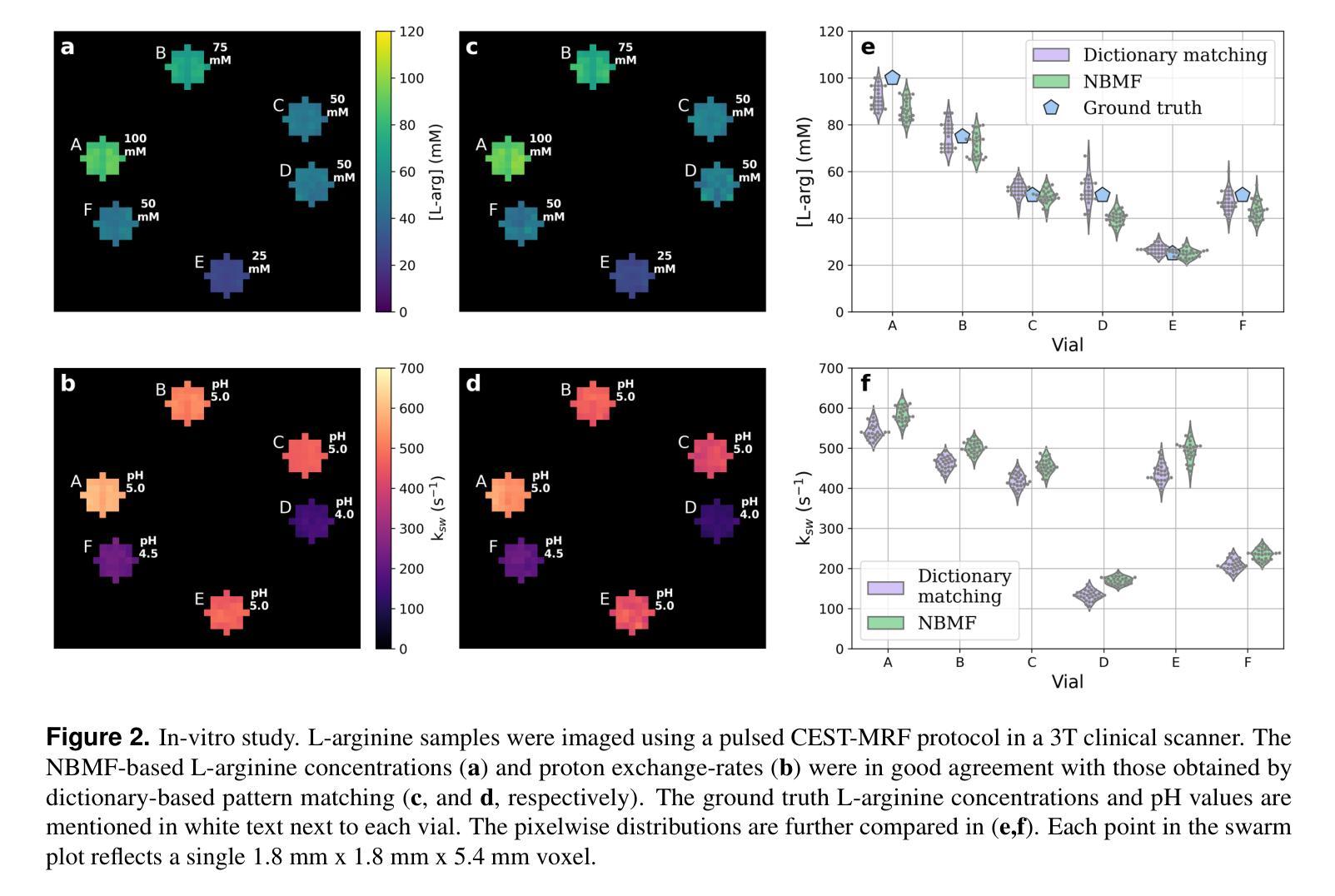
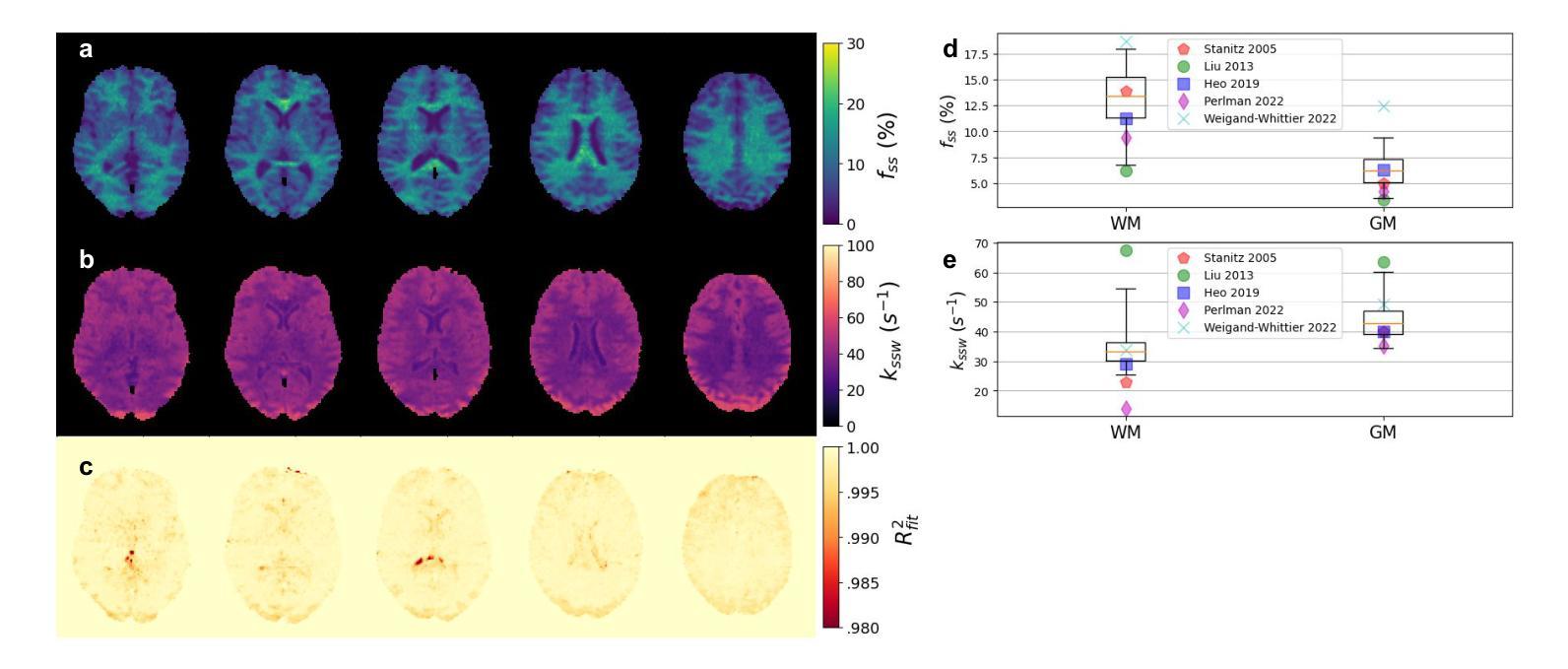
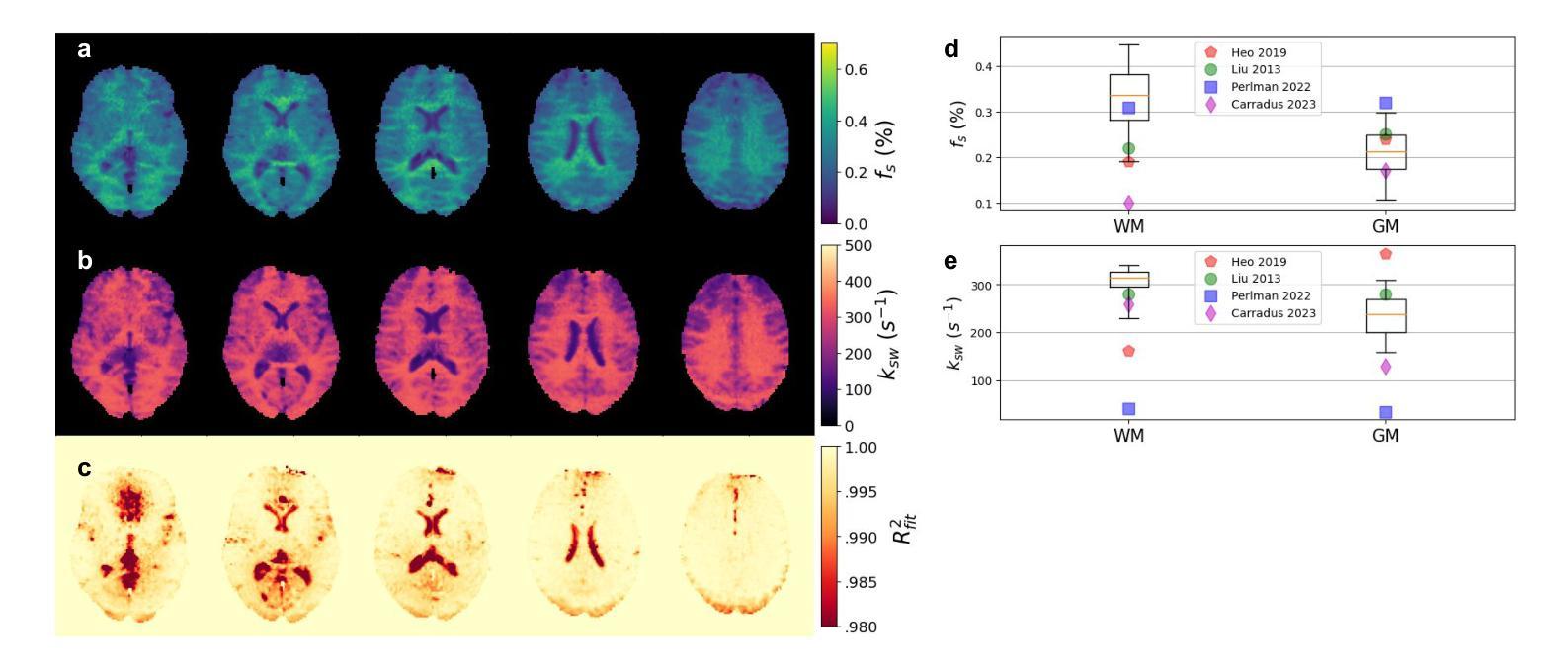
Biophysical model fitting plays a key role in obtaining quantitative parameters from physiological signals and images. However, the model complexity for molecular magnetic resonance imaging (MRI) often translates into excessive computation time, which makes clinical use impractical. Here, we present a generic computational approach for solving the parameter extraction inverse problem posed by ordinary differential equation (ODE) modeling coupled with experimental measurement of the system dynamics. This is achieved by formulating a numerical ODE solver to function as a step-wise analytical one, thereby making it compatible with automatic differentiation-based optimization. This enables efficient gradient-based model fitting, and provides a new approach to parameter quantification based on self-supervised learning from a single data observation. The neural-network-based train-by-fit pipeline was used to quantify semisolid magnetization transfer (MT) and chemical exchange saturation transfer (CEST) amide proton exchange parameters in the human brain, in an in-vivo molecular MRI study (n = 4). The entire pipeline of the first whole brain quantification was completed in 18.3 $\pm$ 8.3 minutes. Reusing the single-subject-trained network for inference in new subjects took 1.0 $\pm$ 0.2 s, to provide results in agreement with literature values and scan-specific fit results.
生物物理模型拟合在从生理信号和图像中获得定量参数方面起着关键作用。然而,分子磁共振成像(MRI)的模型复杂性通常会导致计算时间过长,使得其在临床上的实际应用变得不切实际。在这里,我们提出了一种解决由常微分方程(ODE)建模所构成参数提取反问题的通用计算方法,并结合系统动力学的实验测量。这是通过制定一个数值ODE求解器来作为逐步分析的工具实现的,使其与基于自动微分的优化相兼容。这能够实现高效的基于梯度的模型拟合,并提供一种基于单一数据观察的自我监督学习的新参数量化方法。在活体分子MRI研究(n=4)中,我们利用基于神经网络的拟合训练管道对半固态磁化转移(MT)和化学交换饱和转移(CEST)酰胺质子交换参数进行了量化。首次完成整个大脑量化管道的时间为18.3±8.3分钟。对于新受试者的推理,重用单个受试者训练过的网络需要花费额外的时间和成本进行验证计算并预测后续多个时间点的观测结果仅需耗时为每主体为约一秒钟(每秒传输一帧的速度),并提供与文献值和特定扫描结果相符的结果。这项技术的普及和实用性将在提高计算效率并促进在更高层次的神经成像分析中的进一步应用方面具有重要影响。
论文及项目相关链接
PDF This project was funded by the European Union (ERC, BabyMagnet, project no. 101115639), the Ministry of Innovation, Science and Technology, Israel, and a grant from the Tel Aviv University Center for AI and Data Science (TAD, The Blavatnik AI and Data Science Fund). None of above can be held responsible for views and opinions expressed, which are those of the authors alone
Summary:
生物物理模型拟合在获取生理信号和图像定量参数方面发挥着关键作用。然而,分子磁共振成像(MRI)模型的复杂性往往导致计算时间过长,使其在临床应用中不切实际。本研究提出了一种解决由常微分方程(ODE)建模所引发的参数提取反问题的通用计算方法,该方法结合了系统动力学实验测量。通过构建数值ODE求解器以实现逐步分析,使其与基于自动微分优化兼容。此方法能进行高效梯度基础模型拟合,并提供一种基于自监督学习单一数据观测的参数量化新方法。研究利用神经网络训练管道对半固态磁化转移(MT)和化学交换饱和转移(CEST)酰胺质子交换参数进行量化分析,完成了首个全脑量化流程,耗时约十八分钟。利用单主体训练网络对新主体进行推理分析只需一秒左右,所得结果与文献值和特定扫描结果相符。
Key Takeaways:
- 生物物理模型拟合在生理信号和图像定量参数获取中起关键作用,但分子磁共振成像(MRI)模型计算复杂,临床运用受限。
- 提出一种解决参数提取反问题的通用计算方法,整合常微分方程(ODE)建模和系统动力学实验测量。
- 通过数值ODE求解器模拟逐步分析,兼容自动微分优化,实现高效梯度基础模型拟合。
- 创新性地采用自监督学习方法进行参数量化,基于单一数据观测。
- 应用神经网络训练管道成功量化半固态磁化转移(MT)和化学交换饱和转移(CEST)酰胺质子交换参数。
- 完成首个全脑量化流程,耗时约十八分钟,展示了高效性。
点此查看论文截图